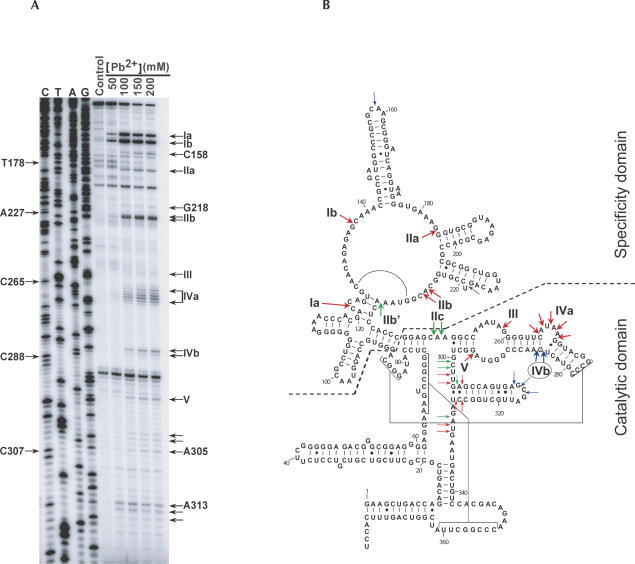
FIGURE 1.
Pb2+ cleavage analysis of M1 RNA in vivo. (A) Total RNA was extracted and used for primer extension analysis with a radiolabeled primer, FP0251. The same primer was also used to generate a DNA sequencing ladder, which in the figure is denoted according to the M1 RNA sequence (C, T, A, G). Reverse transcription products were run on an 8% denaturing gel and subsequently autoradiographed. Pb2+ concentrations used are shown. Control refers to a primer extension performed on RNA from mock-treated cells, to correct for reverse transcription stops due to structure. Site-specific Pb2+-induced cleavage sites are denoted by Roman numerals and are indicated to the right of the autoradiogram. Some additional cleavage sites, along with their assigned nucleotide positions, are indicated as well. (B) The cleavage positions that were reproducibly found in several independent experiments with two different radiolabeled primers (M1–2 and FP0251) are represented on the secondary structure model of M1 RNA (Haas et al. 1996). Red arrows indicate Pb2+ cleavages identified both in vivo and in vitro. Pb2+ cleavages found only either in vivo or in vitro are indicated by blue and green arrows, respectively. The circled site-specific Pb2+-induced cleavage at site IVb is indicative of tRNA substrate binding. Cleavages downstream of C338 were not detectable due to the location of the primer binding sites. Specificity domain and catalytic domain of M1 RNA are indicated on the secondary structure model.