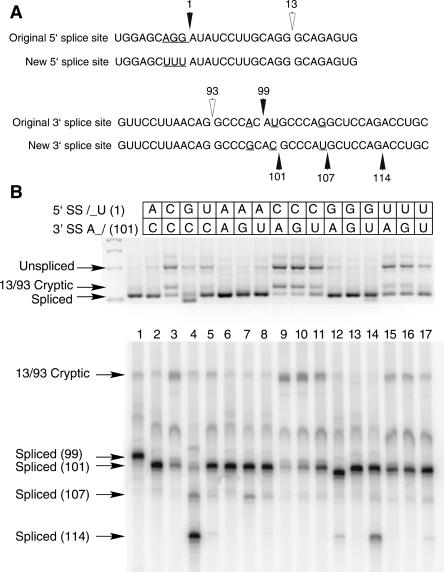
FIGURE 1.
(A) Comparison of the 5′ and 3′ splice site regions of human P120 intron F used for the terminal nucleotide mutant analysis. The top lines show the wild-type genomic sequence and the bottom lines show the sequences used as the starting point for the mutants. The changes were designed to reduce the use of nearby cryptic splice sites and to optimize the branch site to 3′ splice site distance. (B) Pattern of in vivo splicing of the original wild type (lane 1), the new wild type (lane 2), and the 15 possible mutants of the terminal nucleotides as indicated by the key at the top. The indicated minigene constructs were transfected into CHO cells and total RNA prepared after 48 h. The splicing pattern of the P120 intron F was determined by RT-PCR amplification and separation of the products on an agarose gel (top) and a denaturing polyacrylamide gel (bottom). The positions of spliced and unspliced PCR products are shown as well as the position of the U2-dependent 13/93 cryptic splice that is activated in some constructs. The numbers at the left indicate the position of the 3′ splice site. In all cases, except for the 13/93 product, the U12-dependent 5′ splice site at position 1 was used.