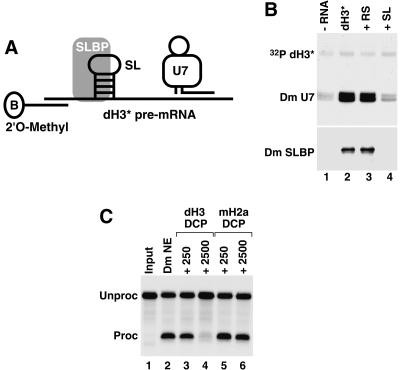
FIGURE 3.
The role of SLBP and the HDE in Drosophila processing. (A) The scheme for purification of processing complexes containing SLBP and U7 snRNP assembled on the dH3 * pre-mRNA. The adapter 2 O-methyl oligonucleotide complementary to the first 17 nt of the pre- ′ mRNA contains biotin (B) on the 3 ′end. (B) Detection of the Drosophila U7 snRNA (Dm U7) by Northern blotting (top) and SLBP (Dm SLBP) by Western blotting (bottom ) in Drosophila processing complexes formed on the dH3 * pre-mRNA in the absence of any competitor RNA (lane 2 ), in the presence of the reverse stem RNA (lane 3), or the stem–loop RNA (lane 4). The background amount of Dm SLBP and Dm U7 snRNA bound to streptavidin beads in the absence of the dH3 *pre-mRNA is shown in lane 1. A small amount of a radioactive dH3 *pre-mRNA ( 32P dH3 *) was added to each processing reaction to monitor the efficiency of isolation of substrate RNA and to control for ethanol precipitation. (C) Processing of the dH3 * pre-mRNA in a Drosophila nuclear extract under control conditions (lane 2 ) or in the presence of a 250 or 2500 M excess of the downstream cleavage product (DCP) from the dH3 pre-mRNA (lanes 3 ,4) or the mouse H2a-614 pre-mRNA (lanes 5,6).