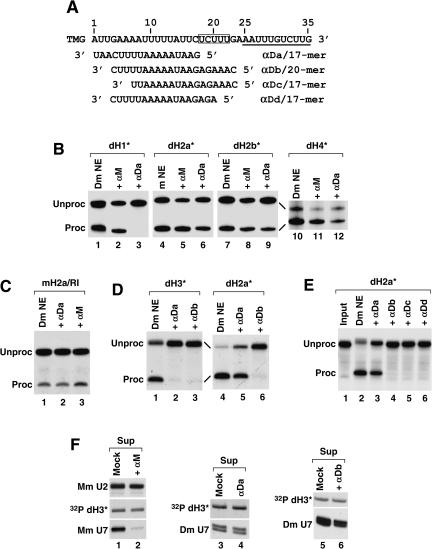
FIGURE 5.
Variable effects of 2′O-methyl oligonucleotides complementary to the Drosophila U7 snRNA on Drosophila histone pre-mRNA processing. (A) The sequence of the first 35 nt of the Drosophila U7 snRNA (written in 5′–3′ orientation) extending from the 5′ terminal tri methyl guanosine cap (TMG) to the end of the Sm binding site (underlined) and the sequences of the four 2′O-methyl oligonucleotides complementary to the 5′ end of the U7 snRNA (written in 3′–5′ orientation). The highly conserved UCUUU sequence in the U7 snRNA is boxed. (B) Processing of the dH1*, dH2a*, dH2b*, and dH4* histone pre-mRNAs in a Drosophila nuclear extract under control processing conditions (lanes 1,4,7,10) or after preincubation of the nuclear extract with either the αM (lanes 2,5,8,11) or the αDa oligonucleotide (lanes 3,6,9,12). (C) Processing of the mouse histone H2a/RI pre-mRNA in a Drosophila nuclear extract under control processing conditions (lane 1) or after preincubation of the nuclear extract with either the αDa (lane 2) or the αM oligonucleotide (lane 3). (D) Processing of the dH3* and dH2a* pre-mRNAs in a Drosophila nuclear extract under control processing conditions (lanes 1,4) or after preincubation of the nuclear extract with either the αDa (lanes 2,5) or the αDb oligonucleotide (lanes 3,6). (E) Processing of the dH2a* pre-mRNA in a Drosophila nuclear extract under control processing conditions (lane 2) or after preincubation of the nuclear extract with the indicated oligonucleotides complementary to the Drosophila U7 snRNA (lanes 3–6). Lane 1 contains the input pre-mRNA. (F) The indicated biotinylated 2′-O-methyl oligonucleotides were incubated with either a mouse (lane 1) or a Drosophila (lanes 4,6) nuclear extract and collected on streptavidin beads. The level of U7 snRNA left in the supernatant was determined by Northern blotting. In the mock experiments (lanes 1,3,5), the nuclear extracts were preincubated with streptavidin beads in the absence of any oligonucleotide. A small amount of the labeled pre-mRNA was added to the extract to control for the efficiency of precipitation (32P dH3*). The mouse U2 snRNA (Mm U2) was detected by Northern blotting to determine the specificity of depletion of the mouse U7 snRNA (Mm U7) in the mouse nuclear extract (lanes 1,2).