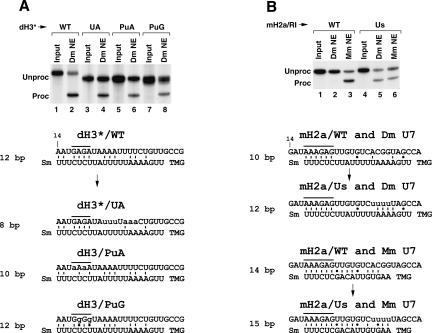
FIGURE 6.
Effects of mutations within the HDEs of the dH3* and H2a/RI pre-mRNAs on 3′ end processing. (A) Processing of the uniformly labeled wild-type dH3* histone pre-mRNA (lanes 1,2) and its mutants, UA, PuA, and PuG (lanes 3–8), in a Drosophila nuclear extract. The sequence of the pre-mRNAs and their likely base-pairing with the Drosophila U7 snRNA are shown at the bottom. GU base pairs are indicated with dots. In each duplex, the pre-mRNA sequence beginning with the 14th nucleotide following the stem–loop is shown at the top and the U7 snRNA sequence is shown at the bottom. The purine core is overlined and the mutated nucleotides are written in lowercase letters. (B) Processing of the wild-type mouse H2a/RI pre-mRNA (lanes 1–3) and the mutant mH2a/Us pre-mRNA containing four Us in the HDE (lanes 4–6) in a Drosophila (lanes 2,5) or a mouse (lanes 3,6) nuclear extract. Likely base-pairing schemes between the pre-mRNAs and either the Drosophila U7 snRNA or mouse U7 snRNA are shown at the bottom.