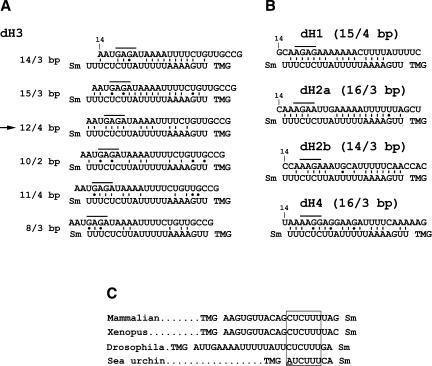
FIGURE 7.
Potential base-pairing schemes between Drosophila pre-mRNAs and the Drosophila U7 snRNA. (A) Several possible base-pairing alignments between the dH3-specific HDE (top sequence) and the first 21 nt of the Drosophila U7 snRNA (bottom sequence). The sequence of the HDE starts 14 nt downstream of the stem–loop (10 nt after the cleavage site). The purine core located between nt 17 and 20 is overlined. The Sm indicates the Sm binding site and the TMG indicates the tri-methyl guanosine cap at the 5′ end of the U7 snRNA. There are two additional nucleotides between the last indicated nucleotide of the U7 snRNA and the Sm binding site. Due to the proximity to the Sm binding site these nucleotides are unlikely to base pair with the pre-mRNA and are not included. The arrow indicates the most favorable alignment that forms a relatively stable duplex and contains the highest number of base pairs between the purine core of the HDE and the UCUUU sequence of the U7 snRNA. (B) The most favorable alignments between the HDE from the four remaining Drosophila histone pre-mRNAs, H1, H2a, H2b, and H4, and the Drosophila U7 snRNA. The numbers in parentheses indicate the total number of base pairs within the duplex and the number of the base pairs formed between the purine core of the HDE and the CUCUUU sequence of the U7 snRNA, respectively. (C) The sequences located between the trimethyl guanosine (TMG) cap and the Sm binding site of the U7 snRNAs from evolutionarily distant organisms. The highly conserved CUCUUU sequence is boxed. The adenosine in the sea urchin U7 snRNA departing from the consensus is underlined.