FIGURE 1.
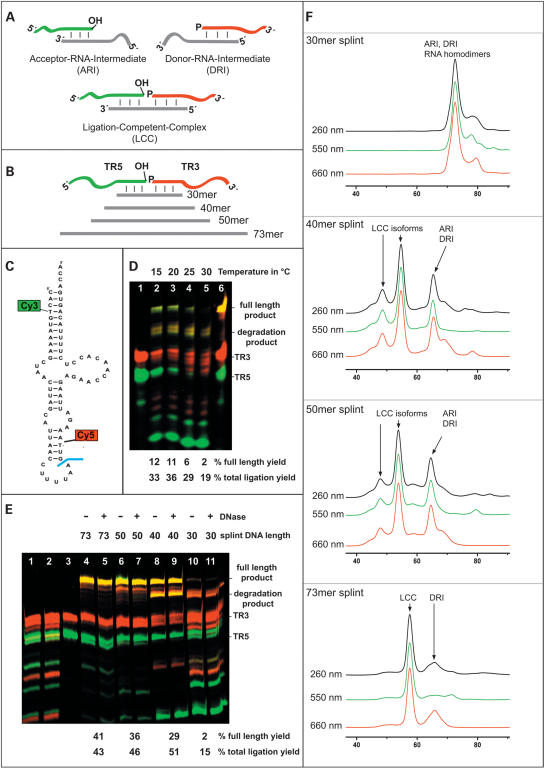
Splint ligation of mitochondrial tRNALys. (A) composition of ARI, DRI, and LCC; phosphates necessary for ligation are indicated by “p,” and hydroxyl groups necessary for ligation are indicated by “OH”.; DNA splints are in light gray; the phosphate acceptor RNA is in green and the donor RNA in red throughout the figure. (B) Splints of 30, 40, 50, and 73 nucleotides and RNA fragments TR5 and TR3. (C) tRNALys with attachment sites for Cy3 and Cy5. The ligation site is indicated in blue. (D) PAGE of ligation reactions mediated by a 30mer splint at different temperatures. (Lane 1) Unligated fragments TR3,TR5. (Lanes 2–5) Ligation reactions at 15° C, 20° C, 25° C, and 30° C. (Lane 6) Contains size markers, TR3, TR5, and full-length product originating from an unrelated synthesis. Total ligation yield and full-length product yield are indicated below the PAGE. (E) Dependence of ligation yield on splint DNA length. (Lane 1) Standard ligation without cDNA template. (Lane 2) Standard ligation to which T4-DNA ligase was added after incubation with DNase. (Lane 3) Unligated fragments TR3, TR5. (Lanes 4–7) Standard ligations with splints of different length. Samples in lanes 5,7,9,11 were incubated with DNase after the ligation reaction. (F) SEC profiles of hybridization mixtures monitored by absorption at different wavelengths. The length of the splint is indicated in each panel. ARIs, DRIs, RNA dimers, LCCs, and structural isoforms as identified by accompanying titration experiments are indicated.