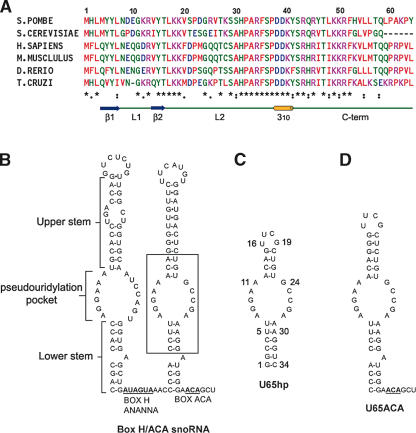
FIGURE 1.
Sequence alignment of Nop10p and secondary structure of snoRNA. (A) Sequence alignments of Nop10. The residues are color coded as follows: red, hydrophobic; blue, negatively charged; pink, positively charged; and green, uncharged polar. The asterisks denote sequence identity, the colon denotes conserved substitutions, and the period denotes semiconserved substitutions. The sequence alignment was done using ClustalW (http://www.ebi.ac.uk/clustalw). A schematic of the secondary structure elements of Nop10p is indicated below the consensus. (B) Secondary structure of human U65snoRNA. (C,D) Sequence and secondary structure of U65hp (C) and U65ACA (D). Other RNAs used in this study include U65s (substrate RNA sequence 5′-UCGGCUCUUCCU), a 16-bp dsRNA capped by an AGAA tetraloop (Wu et al. 2004), and U65 3′ terminal hairpin (9 bp and UUCAUGU loop).