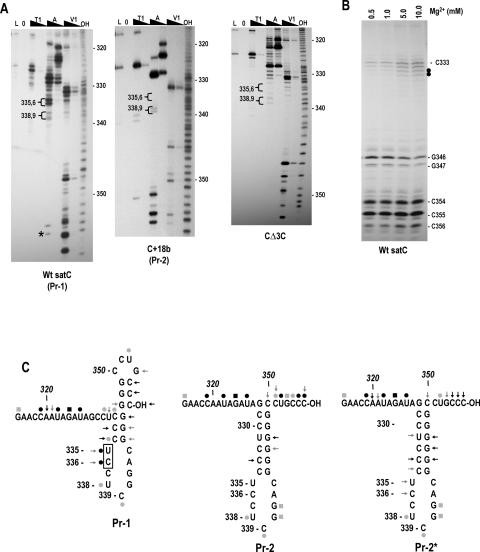
FIGURE 3.
SatC with additional 18 plasmid-derived bases at the 3′ end has a structurally altered Pr region. (A) SatC, C+18b, and CΔ3C transcripts were subjected to partial treatment with two concentrations each of RNase T1 (T1), RNase A (A), or RNase V1 (V1). (L) RNase T1 ladder; (0) no added enzymes; (OH) transcripts were partially digested with alkaline buffer. U335/C336 and U338/C339 are in brackets since their susceptibility to RNase A was always clearly different in Pr-1 and Pr-2 forms of the Pr. Asterisk denotes cleavage was not consistent in wild-type satC transcripts. (B) Wild-type satC 3′ end-labeled transcripts were subjected to RNase V1 digestion with increasing concentrations of Mg2+. Closed circles denote residues whose cleavage was dependent on higher levels of Mg2+. (C) Possible secondary structures of Pr-1, Pr-2, and Pr-2*. (←) RNase V1; (•) RNase A; (▪) RNase T1. Intensity of the symbols implies relative intensity of the cleavages. Boxed residues are cleaved differentially by RNase V1 in the presence of different concentrations of Mg2+. The structure shown for Pr-2 and Pr-2* is the phylogenetically conserved structure for carmoviral Pr. Representative autoradiographs for Pr-2* are shown in Figure 5B ▶.