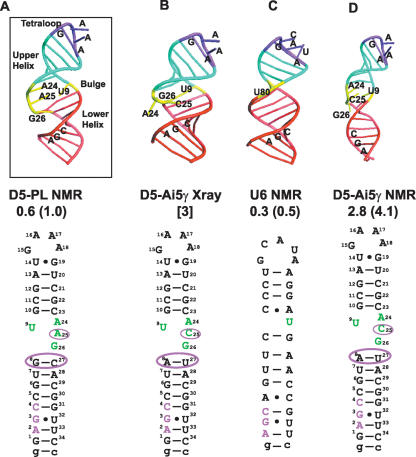
FIGURE 3.
Schematic of D5 and U6 structures highlighting differences and similarities. (A) Solution NMR structure of D5-PL depicting the three critical structural regions of the tetraloop, bulge, and triad AGC regions. (B) X-Ray crystal structure of D5-Ai5γ depicting extra helical conformation of the bulge A24 and C25 nucleotides. (C) Solution NMR structure of U6 ISL depicting the three critical structural regions of the tetraloop, bulge, and triad AGC regions. (D) Solution NMR structure of D5-Ai5γ. The atomic coordinates for the D5-Ai5γ and U6 structures are from PDB codes 1KXK, 1R2P, and 1XHP (6–8). The overall RMSD from the mean structure and the RMS pairwise difference between structures are shown below each structure based on the 10 lowest energy conformers for the NMR structures. For the X-ray structure, we show the resolution. For ease of comparison, we also depict the 2D sequences below each structure.