Abstract
To determine mechanistically how siRNAs mediate transcriptional gene silencing (TGS) in human cells, we have measured histone methylation at targeted promoters, the dependency on active transcription, and whether or not both strands of the siRNA are required for siRNA-mediated TGS. We report here that siRNA treatment increases both H3K9 and H3K27 methylation of the targeted EF1A promoter and that this increase is dependent on nuclear specific delivery of the siRNA. We also find that TGS can be directed by the antisense strand alone, and requires active transcription by RNA polymerase II in human cells as evidenced by sensitivity to α-amanatin. The observation of antisense strand-specific siRNA-mediated TGS of EF1A was substantiated by targeting the U3 region of the HIV-1 LTR/promoter. Furthermore, we show that the antisense strand of siRNA EF52 associates with the transiently expressed Flag-tagged DNMT3A, the targeted EF1A promoter, and trimethylated H3K27. The observations reported here implicate a functional link between siRNA-mediated targeting of genomic regions (promoters), RNA Pol II function, histone methylation, and DNMT3A and support a paradigm in which the antisense strands of siRNAs alone can direct sequence-specific transcriptional gene silencing in human cells.
Keywords: siRNA, RNAi, siRNA mediated TGS, histone methylation, transcriptional gene silencing (TGS), HIV-1, promoter targeting
INTRODUCTION
Transcriptional gene silencing (TGS) caused by small interfering RNAs (siRNA) was first observed in transformed tobacco plants exhibiting a suppressed phenotype of a transgene. Further analysis indicated that DNA methylation of the targeted gene was involved in the suppression (Matzke et al. 1989). TGS mediated by siRNAs was also found in plants infected with a cytoplasmic dsRNA virus; integrated transgenes with promoters homologous to sequences in the virus were found to be silenced (Wassenegger et al. 1994). siRNAs that target promoter sequences have also been shown to cause TGS in Drosophila (Pal-Bhadra et al. 2002). In plants, siRNA-induced silencing of endogenous genes homologous to the transgene is often observed and is accompanied with DNA methylation of homologous sequences. TGS in Arabidopsis thaliana requires siRNA metabolizing factors (Zilberman et al. 2003; Chan et al. 2004), and maintenance of Schizosaccharomyces pombe centromeric heterochromatin depends on siRNA-directed histone 3 lysine 9 (H3K9) methylation (Mette et al. 2000; Jones et al. 2001; Volpe et al. 2002).
Until recently it remained unknown whether siRNA-mediated TGS occurred in mammalian cells. However, recent reports have documented that siRNAs targeted to three different genes at or near promoters, can induce transcriptional silencing (Morris et al. 2004a; Castanotto et al. 2005; Suzuki et al. 2005; Ting et al. 2005). Three of these studies demonstrated that transcriptional inhibition was associated with increased de novo DNA methylation within the siRNA-targeted sequence (Morris et al. 2004a; Castanotto et al. 2005; Suzuki et al. 2005), required nuclear specific delivery (Morris et al. 2004a), and was relieved by treatment with the drugs 5′-azacytidine (5′-AzaC) and trichostatin A (TSA), inhibitors of DNA methylation and histone deacetylation, respectively (Morris et al. 2004a). The observation that TSA was involved in suppressing siRNA-mediated TGS (Morris et al. 2004a) and that others have observed a correlation with siRNA-mediated transcriptional silencing and histone 3 lysine 9 (H3K9) methylation in the absence of DNA methylation (Ting et al. 2005), suggested that chromatin modifications play a more profound role in the observed silencing. Moreover, silencing of genes by both DNA and histone methylation has been shown in cancer cells to depend first on H3K9 methylation (Strunnikova et al. 2005), while in plants, histone 3 lysine 27 (H3K27) trimethylation is also required for the establishment of DNA methylation and gene silencing (Lindroth et al. 2004).
RESULTS AND DISCUSSION
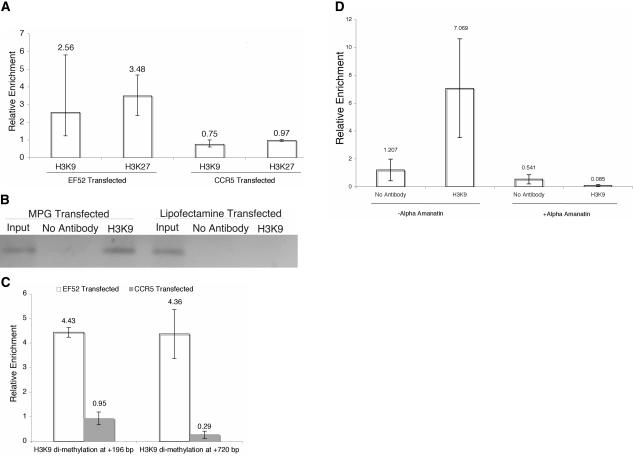
siRNA EF52 is homologous to sequence −104 to −125 relative to the transcriptional start site in the EF1A promoter and was shown previously to mediate TGS of the endogenous EF1A gene (Morris et al. 2004a). To investigate the histone methyl mark induced by siRNA EF52, we transfected 293T cells with either EF52 or the control CCR5 siRNA (Morris et al. 2004a) using the peptide MPG, which transports siRNAs to the nucleus (Morris et al. 1997). EF52-treated cultures exhibited an increase in both H3K9 and H3K27 methylation relative to controls (Fig. 1A ▶). Moreover, the induction of H3K9 methylation was contingent on nuclear-specific delivery of the EF52 siRNA (Fig. 1B ▶). In X inactivation in mammals, silencing is mediated by histone methylation that has been shown to exhibit a histone methyl mark over large distances (Sharp et al. 2002; Danzer and Wallrath 2004). To determine the extent to which the observed histone methyl mark can spread in EF52-treated cultures, we assessed H3K9 dimethylation up to 720 bp downstream of the EF1A transcriptional start site. An approximately fourfold increase in H3K9 methylation was observed for at least 720 bp downstream of the EF1A transcriptional start site in EF52 but not in control transfected cultures (Fig. 1C ▶), similar to observations in siRNA targeting of the CDH1 promoter (Ting et al. 2005).
FIGURE 1.
siRNA-mediated histone methylation and the requirement for RNA polymerase II function. (A) MPG-transfected EF52 siRNA induces histone methylation. Histone 3 lysine 9 (H3K9) dimethylation and histone 3 lysine 27 (H3K27) trimethylation were determined from 293T cells transfected with EF52 or the control CCR5 siRNAs (10 nM) using the nuclear-specific amphipathic peptide MPG (Morris et al. 1997). Forty-eight hours post-transfection, ChIP assays were performed specifically for the EF1A promoter (Morris et al. 2004a). Results represent a minimum of two independent experiments with the range shown. (B) Nuclear-specific delivery is required for histone methylation. MPG and Lipofectamine 2000 transfection reagents were used to transfect EF52-Cy3+ siRNAs into 293T cells. Forty-eight hours following transfection, cultures were collected and a ChIP assay was performed. (C) H3K9 dimethylation can spread 720 bp downstream of the targeted EF1A promoter. H3K9 dimethylation was measured by quantitative PCR either 196 or 720 bp downstream from the targeted EF52 promoter. siRNAs EF52 and CCR5 (10 nM) were transfected with MPG into 293T cells, and ChIP assays were performed 48 h later. Two independent experiments with the range are shown. (D) Treatment of 293T cells with α-amanatin (0.05 μg/mL) 24 h following transfection with siRNA EF52 reduces H3K9 methylation to levels comparable to the no-antibody controls. Results from three independent experiments are shown with the standard deviations.
Recent reports in S. pombe have demonstrated that RNA polymerase II (Pol II) transcription is required for siRNA-mediated TGS. To determine if RNA Pol II transcription is required for siRNA-mediated transcriptional silencing in human cells, we transfected 293T cells with EF52 siRNA using MPG and 24 h later treated half of the cultures with α-amanatin to inhibit RNA Pol II. We then assayed for H3K9 methylation and found that α-amanatin treatment does inhibit EF52 siRNA-mediated H3K9 methylation (Fig. 1D ▶). These data suggest that, similar to S. pombe (Kato et al. 2005), RNA Pol II-mediated transcription is required for siRNA-mediated TGS at some loci in human cells.
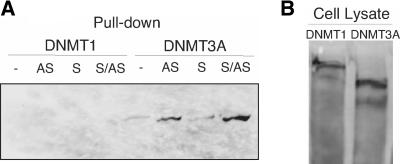
For some promoters, TGS is associated with an increase in DNA methylation (Morris et al. 2004a; Castanotto et al. 2005; Suzuki et al. 2005). We find that TGS is associated with an increase in both H3K9 and H3K27 methylation, and DNA methyltransferases are thought to be part of complexes containing factors that associate with methylated histones (Fuks et al. 2003). In plants, H3K9 and H3K27 methyl marks provide a combinatorial histone code to recruit the DNA methyl-transferase CMT3 to silent loci (Lindroth et al. 2004). Moreover, recombinant, purified mouse DNA methyltransferase 3A (DNMT3A) has been reported to bind double-stranded siRNAs (Jeffery and Nakielny 2004). To extend our understanding of the role that DNA methyltransferases might play in siRNA-mediated TGS in human cells, we developed an siRNA pull-down assay and screened for complexes containing the DNA methyltransferases DNMT1 or DNMT3A. We found that in crude extracts, the antisense strand of EF52 bound to Flag-tagged DNMT3A with similar efficiency as double-stranded siRNA (Fig. 2A ▶). There was no observed interaction with DNMT1 (Fig. 2A ▶), despite similar expression of both Flag-tagged DNMT1 and DNMT3A in the transfected cells (Fig. 2B ▶). These data suggest that the antisense strand may direct the observed siRNA-mediated TGS through interactions with a complex that also contains DNMT3A.
FIGURE 2.
siRNA interactions with DNMT3A. (A) Western blot analysis following biotin/avidin pull-down shows that antisense and dsRNA EF52 associate with DNMT3A but not DNMT1. Flag-tagged DNMT1 and DNMT3A transfected 293T cell lysates were generated and incubated with a total of 500 nM biotin-labeled sense (S), anti-sense (AS), or double-stranded (S/AS) siRNA, and the control dsRNA without a biotin label (−). Immunostaining for the Western blot was performed with an anti-Flag antibody. (B) Western blot analysis showing comparable expression levels of both Flag-tagged DNMT1 and DNMT3A in the transfected 293T cell lysates.
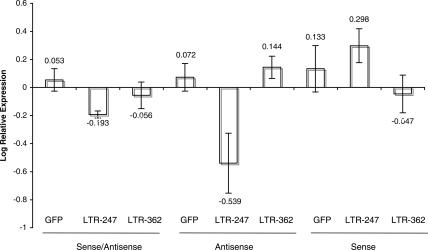
The observation that the antisense strand of the siRNA is bound to DNMT3A in the cell lysates containing Flag-tagged DNMT3A prompted us to test whether the antisense strand might function alone to direct TGS. We designed plasmids containing the U6 promoter to express the antisense, sense, or combined dsRNA targeted to two regions of the U3 in the HIV-1 LTR/promoter (LTR-247 and 362, respectively) (Table 1 ▶). These plasmids and an HIV-1 tat expression plasmid were cotransfected into 1G5 cells which contain an integrated HIV-1 LTR that can express firefly luciferase with an SV40 poly(A) in place of the 3′-LTR (Aguilar-Cordova et al. 1994) when HIV-1 Tat is present. We found that the antisense LTR-247 siRNA produced TGS in 1G5 cells at levels comparable to the dsRNA transfected samples (Fig. 3 ▶). In other experiments, we found that the antisense LTR-247 was also capable of directing H3K27 methylation in an RNA Pol II-dependent manner (data not shown).
TABLE 1.
siRNAs and respective siRNA target sequence used in the HIV-1 U3 targeting
| siRNA (Position) | Sequence (Target) | % GC |
| 247 (249–267 in LTR of HIV subtype B) | GTGTTAGAGTGGAGGTTTG | 47.4 |
| 362 (354–372 in LTR of HIV subtype B) | CTTTCCGCTGGGGACTTTC | 57.9 |
| GFP (108–126 in GFP mRNA) | CGATGCCACCTACGGCAAG | 63.2 |
| R5 Control (787–805 in CCR5 mRNA) | TTCTTTGGCCTGAATAATT | 31.6 |
FIGURE 3.
LTR-247 sense/antisense and antisense alone are capable of targeting the U3 region of the HIV-1 LTR and inhibiting Tat-mediated expression of fire fly luciferase in 1G5 cells. Results are from four independent experiments measured 24 h post-transfection with log relative expression and standard error of the means shown. Values represent luciferase expression relative to the experimental mean.
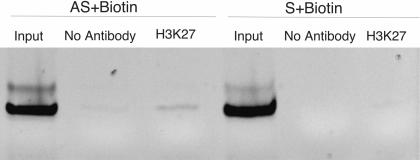
The observations that (1) siRNA-mediated TGS is reversible with the addition of both 5-AzaC and TSA (Morris et al. 2004a); (2) siRNA EF52 associates with Flag-tagged DNMT3A in cell extracts (Fig. 2B ▶; Jeffery and Nakielny 2004); and (3) histone 3 lysine methylation, both K9 and K27, is present at the targeted promoter (Fig. 1A–D ▶), are consistent with a complex forming at the promoter during the establishment of TGS. To investigate this putative complex further, we performed a triple-pull-down assay. 293T cells were transfected with Flag-tagged DNMT3A and 24 h later transfected again with biotin labeled either sense or antisense EF52 siRNAs using MPG. Forty-eight hours later a ChIP for H3K27 was performed, followed by a Flag-tagged DNMT3A immunoprecipitation and then a siRNA biotin/avidin pull-down and PCR for the targeted promoter in the final elute. We found that the antisense EF52 strand was enriched twofold and 3.5-fold relative to the no-antibody and sense-alone controls, respectively (Fig. 4 ▶).
FIGURE 4.
Biotin-labeled antisense siRNA EF52 (AS+Biotin) coprecipitates with trimethylated H3K27, Flag-tagged DNMT3A, and the targeted EF1A promoter, while the sense EF52 (S+Biotin) does not. Results are representative of two independent experiments.
The initial discovery that promoter-targeted siRNAs can cause gene silencing in human cells demonstrated that small RNAs in mammals, Drosophila, Caenorhabditis elegans, and plants can regulate gene expression by three conserved mechanisms: transcriptional gene silencing (TGS), post-transcriptional gene silencing (PTGS), and translational inhibition (microRNA inhibition) (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). While there are many functional similarities between siRNA-mediated TGS in mammals, S. pombe, and plants, the underlying mechanism(s) may be somewhat varied. The observation that the antisense strand of EF52 coimmunoprecipitates in vivo with transiently expressed Flag-tagged DNMT3A, tri-methylated H3K27, and the targeted promoter as well as the observation that the antisense LTR-247 is efficacious in suppressing HIV-1 Tat-induced U3-mediated transcription suggest a mechanism of action. One model might be that the antisense strand of the siRNA is transferred in the nucleus to a complex that also contains DNMT3A. This complex might then be guided by RNA homology to the promoter and direct histone methylation as the primary silencing event that with time may lead to increased DNA methylation. H3K9 and/or H3K27 methylation correlates well with a transcriptionally silent state of chromatin (Nguyen et al. 2002; Mutskov and Felsenfeld 2004), and we observed H3K9 and H3K27 methylation resulting from either promoter-targeted double-stranded siRNA or the antisense siRNA treatment. Interestingly, DNMT3A has been shown to strongly associate with HDAC-1 and Suv39H1 (Fuks et al. 2001, 2003; Datta et al. 2003), proteins known to be associated with silent chromatin. Whether such a large complex with these proteins is involved in the observed antisense-mediated transcriptional silencing shown here remains to be determined.
Using human cells, we show here that EF1A and HIV-1 promoter-directed siRNAs, even just the antisense strand, can mediate transcriptional silencing and increase histone methylation. We also demonstrate that the antisense strand of the EF52 siRNA coprecipitates with DNMT3A, H3K27 trimethylated histones, and the targeted promoter in vivo, whereas the sense strand alone did not. Importantly, the observation that siRNAs direct histone methylation and that this effect is prevented by the inhibition of RNA Pol II suggests that transcription is required for siRNA-mediated TGS as well as that siRNAs, specifically the antisense strand, may function to direct and/or write the histone code. Taken together these data propose that the antisense strand of promoter-directed siRNAs mediates control of gene expression in an RNA Pol II-dependent fashion through epigenetic modifications first involving histone 3 methylation. These findings suggest that the antisense strand of the promoter-directed siRNAs play a pivotal and underappreciated role in gene regulation that could be conceptualized for therapeutic applications in humans.
MATERIALS AND METHODS
Chromatin immunoprecipitation assay (ChIP)
Chromatin immunoprecipitation was performed on 4.0 × 106 293T cells transfected with siRNA EF52 or control CCR5 (Ambion Silencer, 10 nM final concentration) using MPG at a 10:1 charge ratio (MPG:siRNA) as described in Morris et al. (1997, 2004a). Forty-eight hours following transfection, cultures were collected and ChIP assays were performed as described (Strahl-Bolsinger et al. 1997). Briefly, the treated cultures were formaldehyde cross-linked (1%, 10 min, room temperature) and then stopped by adding glycine at a final concentration (0.125 M, 10 min, room temperature). The cells were then washed twice in 1 × PBS + 1/1000 PMSF (stock PMSF at 0.5 M), resuspended in 600 μL of ChIP lysis buffer (50 mM HEPES at pH 7.5, 140 mM NaCl, 10% Triton X-100, 0.1% NaDeoxycholate, 1/1000 PMSF) and incubated (10 min on ice). Next, the samples were centrifuged (5000 rpm, 5 min, 4°C), resuspended in 600 μl of ChIP lysis buffer, incubated (10 min on ice), and then sonicated (Branson 50 cell machine, six intervals with 20-sec pulses and 2-min rests). The sonicated samples were then centrifuged (14,000 rpm, 10 min, 4°C) and the supernatants were removed and pre-cleared with 30 μL of protein A/salmon sperm (Upstate catalog #16-157; 15 min, 4°C, rotating platform). The pre-cleared supernatants were then centrifuged (14,000 rpm, 5 min, 4°C), and the supernatants were removed with equivalent aliquots (from the pre-cleared supernatants) used. The samples consisted of the no-antibody control, anti-dimethyl-H3K9 pull-down (H3K9), and/or anti-trimethyl-H3K27 pull-down (H3K27) and were incubated either without an antibody (no antibody control), or for H3K9 samples, the anti-dimethyl-Histone H3 (Lys 9) antibody (Upstate catalog #07-441) was used; and for H3K27 samples, the anti-trimethyl-Histone H3 (Lys 27; Upstate catalog #07-449) was used (3 h to overnight, 4°C, rotating platform). The samples were then treated with 10 μL of protein A/salmon sperm (Upstate; 15 min, room temperature, rotating platform) and then pulled down (10,000 rpm, 1 min, 4°C) and washed. The no-antibody control supernatants were saved and used as input controls. The washes consisted of two washes with 1 mL of ChIP lysis buffer, two washes with 1 mL of ChIP lysis buffer high salt (50 mM HEPES at pH 7.5, 500 mM NaCl, 1% Triton X-100, 0.1% NaDeoxycholate, 1/1000 PMSF), followed by two washes with 1 mL of ChIP wash buffer (10 mM Tris at pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% NaDeoxycholate, 1 mM EDTA). For each wash, the samples were incubated (3 min, room temperature, rotating platform), followed by centrifugation (14,000 rpm, 3 min, room temperature). After the final wash, the complexes were eluted by two treatments of 100 μL of elution buffer (50 mM Tris at pH 8.0, 1% SDS, 10 mM EDTA) for 10 min at 65°C, followed by centrifugation (14,000 rpm, 3 min, room temperature). The eluted complexes along with the initial aliquot used in the no-antibody control (200 μL) were then reverse cross-linked by adding 1 μL of RNase A (10 mg/mL) and 20 μL of 5 M NaCl to each sample, and incubated (4–6 h, 65°C). The reverse cross-linked samples were then treated with 10 μL of 0.5M EDTA, 20 μL of 1 M Tris-HCl at pH 6.5, and 2 μL of 10 mg/mL Proteinase K (1 h, 45°C) and the DNA was recovered by phenol/chloroform extraction and assayed using PCR for 30 cycles of 94°:55°:72°C at 30:30:30 sec with primers 803 and 804, which specifically overlap the targeted EF1 α promoter (Morris et al. 2004a). The resultant PCR products were quantitated using the integrated density values (IDV) as determined from analysis with an α Innotech gel imager or with quantitative PCR (qPCR); 35 cycles of 94°:55°:72°C at 30:30:30 sec with primers 803 and 804 and standardized to plasmid pVEGFPwP (Morris et al. 2004a,b). To determine the extent of H3K9 spreading at 196 bp downstream of the EF1A transcription start site, primers 783 (5′-GTGGGTGGGAGAGTTCGAG-3′) and 785 (5′-GCGTCGCAGCAGGTCA-3′) were used; and to measure H3K9 methylation at 720 bp downstream of the EF1A transcription start site, primers 775 (5′-GGCGCCGTCCAGGCACCTCG-3′) and 776 (5′-CCAAGATCCAAACTCAA-3′) were used. The HIV-LTR ChIP assays involved a final PCR amplification step with the elutes as template of 28 cycles of 94°:55°:72°C at 15:15:30 sec using primers 5′-247-1 (5′-CCCTGTGAGCCTGCATGGAATGGATGACCC-3′) and 3′-362-2 (5′-CCAGAGAGACCCAGTACAGGCAAAAAGCAG-3′) and visualized on the α Innotech gel imager.
Detection of Flag-tagged proteins in biotin-labeled siRNA pull-downs
A total of 4.0 × 106 293T cells were transfected with 15 μg expression vectors for Flag-tagged DNMT1 or DNMT3A using Lipofectamine 2000. All DNMTs were supplied by A.D. Riggs (Beckman Research Institute of City of Hope [BRICOH]). Forty-eight hours later, the cell lysates (cytoplasmic and nuclear fractions) were isolated each in 500 μL of lysis buffer; cytoplasmic fraction (1 mM PMSF, 20 units RNasin, 10 mM Tris-Cl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40) and nuclear fraction (1 mM PMSF, 20 units RNasin, 20 mM Hepes [pH 7.0], 150 mM NaCl, 2 mM MgCl2, 1 mM DTT, and 0.5% NP-40). A mixture of 250 μL of cytoplasmic and nuclear fractions was incubated for 3 h at 4°C with 500 nM 5′-biotin end-labeled EF52 (BRI/COH DNA/RNA/Peptide Core Facility). Next, Dynal Avidin/magnetic beads (7 × 107 beads) were washed in lysate buffer and then added directly to the siRNA/Flag-tagged containing lysates and incubated at 4°C for 1 h. The siRNA/Flag complexes were pulled down by magnetic bead binding and washed five times in lysate buffer. Finally, the bound protein complexes were eluted from the Avidin-biotin bound beads/siRNA by incubation in 100 μL of elution buffer (50 mM Tris-Cl at pH 6.0, 1 mM EDTA, 2.0 M NaCl, 0.5 M MgCl2) at 65°C for 5 min. The eluted complexes were electrophoresed in SDS-PAGE and subjected to Western blot analysis with an anti-Flag antibody.
To determine specificity of siRNA binding, Flag-tagged DNMT1 and DNMT3A were generated from transfected 293T cells as described above. The cell lysates containing Flag-tagged DNMTs were then incubated with either the siRNA EF52, sense (S) or antisense (AS) EF52 (500 nM) 5′-Biotin end-labeled for 3 h at 4°C. The resultant complexes bound to either S or AS or S/AS (dsRNA) siRNA EF52 were isolated by Dynal Avidin/magnetic beads (7 × 107 beads), washed five times in lysate buffer, and eluted, and protein complexes were electrophoresed in SDS-PAGE and subjected to Western blot analysis with an anti-Flag antibody.
HIV-1 U3-specific LTR targeting
HIV-1 U3 LTR-targeted siRNAs were constructed and cloned into pCR4-TOPO (Invitrogen) by PCR using the 5′ U6+1 as described (Lee et al. 2002) with 3′ primers:
LTR-247(S), 5′-AAAAAAAAGTGTTAGAGTGGAGGTTTGCGGTGTTTCGTCCTTTCCACAA-3′;
LTR-247(AS), 5′-AAAAAAAACAAACCTCCACTCTAACACGGTGTTTCGTCCTTTCCACAA-3′;
LTR-362(S), 5′-AAAAAAAACTTTCCGCTGGGGACTTTCGGTGTTTCGTCCTTTCCACAA-3′;
LTR-362(AS), 5′-AAAAAAAAGAAAGTCCCCAGCGGAAAGCGGTGTTTCGTCCTTTCCACAA-3′;
GFP(S), 5′-AAAAAAAACTTGCCGTAGGTGGCATCGCGGTGTTTCGTCCTTTCCACAA-3′; and
GFP(AS), 5′-AAAAAAAACGATGCCACCTACGGCAAGCGGTGTTTCGTCCTTTCCACAA-3′.
The resultant clones were sequenced and plasmids were cotransfected (1:1 ratio, 0.5 to 0.5 μg) with the HIV-1 Tat expression plasmid pTatdsRed2 (Unwalla et al. 2004) into 2 × 106 1G5 Jurkat cells using the Amaxa Nucleofector. The 1G5 cells were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID (Platt et al. 1998; Wei et al. 2002). Twenty-four hours later luciferase expression was determined using the Dual-Luciferase Reporter Assay System (Promega) and a Veritas microplate luminometer from Turner Biosystems following the manufacturers’ protocols and luciferase expression standardized to living cells.
α-Amanatin-mediated suppression of siRNA-induced TGS
Chromatin immunoprecipitation was performed on 4.0 × 106 293T cells transfected with siRNA EF52 and control CCR5 (10 nM final concentration using MPG as described above) (Morris et al. 1997, 2004a) or on 4 × 106 TZM-Bl cells obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID (Platt et al. 1998; Wei et al. 2002) cotransfected with plasmids pTatdsRed2 (0.5 μg) (33) and p-LTR-247(AS) (0.5 μg). Twenty-four hours following transfection, one-half of the cultures were exposed to α-amanatin (0.05 μg/mL), and 24 h later both the treated and untreated cultures were collected and a ChIP assay was performed as described previously (Strahl-Bolsinger et al. 1997). Cultures were immunoprecipitated with anti-dimethyl-Histone H3 (Lys 9) or anti-trimethyl-Histone (Lys 27), and the final elutes were assayed using PCR for 30–35 cycles (α innotech or qPCR, respectively) of 94°:55°:72°C and 30, 30, and 30 sec, respectively, with primers 803 and 804 (EF52 specific) (Morris et al. 2004a) or 28 cycles with the U3 HIV-1 LTR-specific 5′-247-1 and 3′-362-2 (described previously).
Triple immunoprecipitation: H3K27 ChIP/-Flag-DNMT3A/biotin-RNA
A total of 4.0 × 106 293T cells were plated and 24 h later transfected with Flag-DNMT3A (15 μg). The next day the Flag-DNMT3A-transfected cultures were transfected again with 100 nM EF52 biotin-labeled siRNA (antisense or sense alone) using stock 3.4 μM MPG (3 μL/mL of media). Forty-eight hours following the sense or antisense siRNA/MPG transfection, cultures were collected and a ChIP assay was performed as described previously with a slight modification. Following the immunoprecipitation with the H3K27 trimethyl-specific antibody (1 μg), the elutes were incubated overnight with 40 μL of EZVIEW Red anti-Flag M2 affinity gel beads (Sigma). Next the bound beads were washed three times with TBS-Mod Buffer (50 mM Tris HCl, 400 mM NaCl at pH 7.4) and then eluted by competition with 3× Flag-peptide (15 μg). The resultant elutes were then transferred to 100 μL (6–7 × 108 beads/mL) of Dynabeads M-280 Streptaviden pre-washed in 2× wash buffer (50 mM Tris HCl, 400 mM NaCl at pH 7.4). The elute/bead slurry was incubated at 4°C for 15 min on an orbital shaker followed by capture with a magnetic bead separator. The captured beads were washed three times in 2× wash buffer and then eluted in 100 μL of 2× elute buffer (10 mM Tris-HCL at pH 6.0, 1 mM EDTA, 2.0 M NaCl) at 65°C for 5 min. The resultant elutes were then reverse cross-linked and DNA was recovered by phenol/chloroform extraction followed by PCR for 30–35 cycles of 94°:55°:72°C for the EF1 α promoter with primers 803 and 804 (described previously).
Acknowledgments
We thank R. Allshire, S. Grewal, and A. Verdel for their comments on siRNA-mediated TGS, and J. Longmate for statistical advice. This work was supported by funds from NIH HLB RO1 HL07470 to J.J.R., R01 HL83473 to K.V.M., and NCI Comprehensive Cancer Center Grant 5P30 CA33572 to J.J.R. and K.V.M.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2235106.
REFERENCES
- Aguilar-Cordova, E., Chinen, J., Donehower, L., Lewis, D.E., and Belmont, J.W. 1994. A sensitive reporter cell line for HIV-1 tat activity, HIV-1 inhibitors, and T cell activation effects. AIDS Res. Hum. Retroviruses 10: 295–301. [DOI] [PubMed] [Google Scholar]
- Castanotto, D., Tommasi, S., Li, M., Li, H., Yanow, S., Pfeifer, G.P., and Rossi, J.J. 2005. Short hairpin RNA-directed cytosine (CpG) methylation of the RASSF1A gene promoter in HeLa cells. Mol. Ther. 12: 179–183. [DOI] [PubMed] [Google Scholar]
- Chan, S.W., Zilberman, D., Xie, Z., Johansen, L.K., Carrington, J.C., and Jacobsen, S.E. 2004. RNA silencing genes control de novo DNA methylation. Science 303: 1336. [DOI] [PubMed] [Google Scholar]
- Danzer, J.R. and Wallrath, L.L. 2004. Mechanisms of HP1-mediated gene silencing in Drosophila. Development 131: 3571–3580. [DOI] [PubMed] [Google Scholar]
- Datta, J., Ghoshal, K., Sharma, S.M., Tajima, S., and Jacob, S.T. 2003. Biochemical fractionation reveals association of DNA methyltransferase (Dnmt) 3b with Dnmt1 and that of Dnmt 3a with a histone H3 methyltransferase and Hdac1. J. Cell Biochem. 88: 855–864. [DOI] [PubMed] [Google Scholar]
- Fuks, F., Burgers, W.A., Godin, N., Kasai, M., and Kouzarides, T. 2001. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 20: 2536–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks, F., Hurd, P.J., Deplus, R., and Kouzarides, T. 2003. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 31: 2305–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery, L. and Nakielny, S. 2004. Components of the DNA methylation system of chromatin control are RNA-binding proteins. J. Biol. Chem. 279: 49479–49487. [DOI] [PubMed] [Google Scholar]
- Jones, L., Ratcliff, F., and Baulcombe, D.C. 2001. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr. Biol. 11: 747–757. [DOI] [PubMed] [Google Scholar]
- Kato, H., Goto, D.B., Martienssen, R.A., Urano, T., Furukawa, K., and Murakami, Y. 2005. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 309: 467–469. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–857. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Lee, R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 94: 862–864. [DOI] [PubMed] [Google Scholar]
- Lee, N.S., Dohjima, T., Bauer, G., Li, H., Li, M., Ehsani, A., Salvaterra, P., and Rossi, J. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotech. 19: 500–505. [DOI] [PubMed] [Google Scholar]
- Lindroth, A.M., Shultis, D., Jasencakova, Z., Fuchs, J., Johnson, L., Schubert, D., Patnaik, D., Pradhan, S., Goodrich, J., Schubert, I., et al. 2004. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 23: 4286–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke, M.A., Primig, M., Trnovsky, J., and Matzke, A.J.M. 1989. Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J. 8: 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette, M.F., Aufsatz, W., Van der Winden, J., Matzke, A.J.M., and Matzke, M.A. 2000. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19: 5194–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, M.C., Vidal, P., Chaloin, L., Heitz, F., and Divita, G. 1997. A new peptide vector for efficient delivery of oligonucleotides into mammalian cells. Nucleic Acids Res. 25: 2730–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, K.V., Chan, S.W., Jacobsen, S.E., and Looney, D.J. 2004a. Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305: 1289–1292. [DOI] [PubMed] [Google Scholar]
- Morris, K.V., Gilbert, J., Wong-Staal, F., Gasmi, M., and Looney, D.J. 2004b. Transduction of cell lines and primary cells by FIV-packaged HIV vectors. Mol. Ther. 10: 181–190. [DOI] [PubMed] [Google Scholar]
- Mutskov, V. and Felsenfeld, G. 2004. Silencing of transgene transcription precedes methylation of promoter DNA and histone H3 lysine 9. EMBO J. 23: 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, C.T., Weisenberger, D.J., Velicescu, M., Gonzales, F.A., Lin, J.C., Liang, G., and Jones, P.A. 2002. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 62: 6456–6461. [PubMed] [Google Scholar]
- Pal-Bhadra, M., Bhadra, U., and Birchler, J.A. 2002. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell 9: 315–327. [DOI] [PubMed] [Google Scholar]
- Platt, E.J., Wehrly, K., Kuhmann, S.E., Chesebro, B., and Kabat, D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72: 2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger, S., Hecht, A., Luo, K., and Grunstein, M. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 11: 83–93. [DOI] [PubMed] [Google Scholar]
- Strunnikova, M., Schagdarsurengin, U., Kehlen, A., Garbe, J.C., Stampfer, M.R., and Dammann, R. 2005. Chromatin inactivation precedes de novo DNA methylation during the progressive epigenetic silencing of the RASSF1A promoter. Mol. Cell. Biol. 25: 3923–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., Shijuuku, T., Fukamachi, T., Zaunders, J., Guillemin, G., Cooper, D., and Kelleher, A. 2005. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. J. RNAi Gene Silencing 1: 66–78. [PMC free article] [PubMed] [Google Scholar]
- Ting, A.H., Schuebel, K.E., Herman, J.G., and Baylin, S.B. 2005. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat. Genet. 37: 906–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwalla, H.J., Li, M.J., Kim, J.D., Li, H.T., Ehsani, A., Alluin, J., and Rossi, J.J. 2004. Negative feedback inhibition of HIV-1 by TAT-inducible expression of siRNA. Nat. Biotechnol. 22: 1573–1578. [DOI] [PubMed] [Google Scholar]
- Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I.S., and Martienssen, R.A. 2002. Regulation of heterchromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Wassenegger, M., Graham, M.W., and Wang, M.D. 1994. RNA-directed de novo methylation of genomic sequences in plants. Cell 76: 567–576. [DOI] [PubMed] [Google Scholar]
- Wei, X., Decker, J.M., Liu, H., Zhang, Z., Arani, R.B., Kilby, J.M., Saag, M.S., Wu, X., Shaw, G.M., and Kappes, J.C. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46: 1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., and Jacobsen, S.E. 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719. [DOI] [PubMed] [Google Scholar]