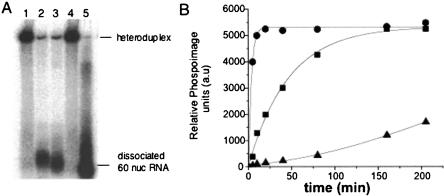
FIGURE 3.
RNA helix destabilizing activity of hantavirus N and HIV-1 Gag. (A) A 60-nt-long oligoribonucleotide, labeled with α-32P-CTP was annealed to an RNA of ~2 kb. The resulting heteroduplex features a central base-paired region 40 nt in length and, for the short RNA, flanking nucleotides in single-stranded configuration. Dissociation of the two RNA would result in liberation of the 60-nt RNA from the heteroduplex. Samples were incubated as described in Materials and Methods for a total of 30 min. Lane 1, RNA heteroduplex without added protein; lane 2, RNA heteroduplex incubated with SNV N; lane 3, RNA heteroduplex incubated with HIV-1 Gag; lane 4, incubated with BSA; lane 5, the 60-nt-long oligoribonucleotide. Samples were incubated with 1:1 molar ration of Hetero duplex RNA:SNV N/HIV 1 Gag. Similar results were obtained in three or more separate experiments. (B) Kinetics of N-mediated heteroduplex dissociation using varying N:S segment RNA ratios. Samples were collected from a helix destabilization assay at the indicated times after initiation of the reaction and the dissociation of the heteroduplex was detected by gel analysis. (•) N:RNA = 6:1, (▪) N:RNA = 1:1, (▵) N:RNA = 1:6. The amount of released labeled RNA was quantified by phosphorimaging. Similar results were obtained from more than three separate experiments.