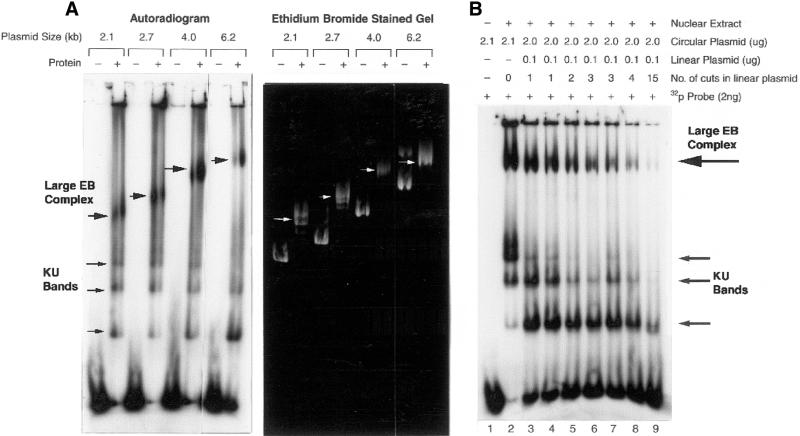
Figure 2.
Binding of radiolabeled probe to the complex comigrates with plasmid DNA and formation of the complex requires Ku-bound DNA ends. (A) Agarose mobility shift assays with or without M059K nuclear extract (0.5 µg) were performed as described in Figure 1. Mobility shift reactions contained closed circular plasmids of increasing molecular size as indicated above the lanes in the autoradiogram and ethidium bromide stained gel. White arrows indicate the position of the large DNA end-binding (EB) complex in the ethidium bromide stained gel. (B) The dependence of probe binding to closed circular pUC18 on DNA ends was determined by competition with unlabeled ends. Lane 1, closed circular pUC18 and a 32P-labeled 144 bp probe without nuclear extract; lane 2, labeled probe, pUC18, and M059K nuclear extract (0.5 µg). Lanes 3–9, competition agarose mobility shift assays containing fixed concentrations of 32P-labeled probe and total DNA, but with increasing concentrations of unlabeled DNA ends previously obtained by digesting pUC18 with restriction enzymes: EcoRI (lane 3), HindIII (lane 4), PvuII (lane 5), RsaI (lane 6), HaeII (lane 7), TaqI (lane 8) and AluI (lane 9). The number of restriction cuts in each reaction is indicated. Upper arrow, the position of the pUC18 end-binding complex; lower arrows, the position of the Ku-bound probe.