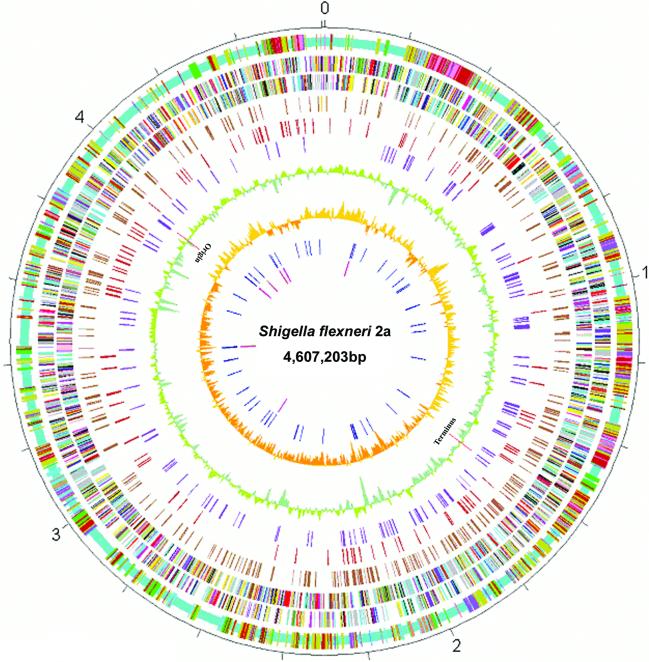
Figure 1.
Circular map of Sf301 chromosome compared with those of E.coli K12 MG1655 and 0157 EDL933. Outer scale is marked in 200 kb. Circles range from 1 (outer circle) to 10 (inner circle). Circle 1, shared collinear backbone (light blue), and Shigella islands (SI) (cyan), K12 islands (KI) (green) and 0157 islands (OI) (tan); co-localized SI and KI (dark salmon); co-localized SI and OI (purple); co-localized KI and OI (dark blue); and co-localized SI, KI and OI (deep pink). Circles 2 and 3, ORFs encoded by leading and lagging strands with color code for functions: salmon, translation, ribosomal structure and biogenesis; light blue, transcription; cyan, DNA replication, recombination and repair; turquoise, cell division; deep pink, posttranslational modification, protein turnover and chaperones; olive drab, cell envelope biogenesis; purple, cell motility and secretion; forest green, inorganic ion transport and metabolism; magenta, signal transduction; red, energy production; sienna, carbohydrate transport and metabolism; yellow, amino acid transport; orange, nucleotide transport and metabolism; gold, co-enzyme transport and metabolism; dark blue, lipid metabolism; blue, secondary metabolites, transport and catabolism; gray, general function prediction only; black, function unclassified or unknown. Circle 4, distribution of pseudogenes. Circles 5 and 6, distribution of IS1 and other IS elements, respectively. Circle 7, G + C content with a window size of 10 kb. Circle 8, GC bias (G – C/G + C). Circle 9, distribution of tRNA genes. Circle 10, distribution of rrn operons. The replication origin and terminus are indicated.