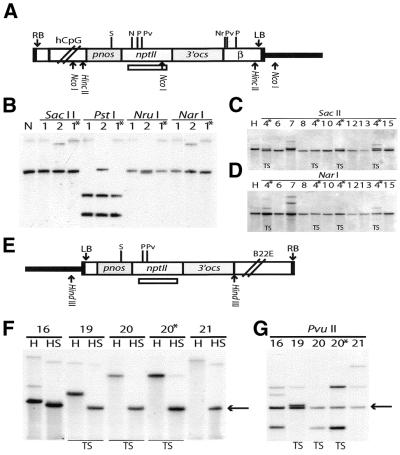
Figure 5.
Methylation analysis of sites in the pnos–nptII region in CpG and Pl:CpG lines. DNA from transgenic plants generated with the constructs ex1 and ex2–4 (A) and plant:CpG-rich (E) was digested with a methylation- sensitive enzyme in combination with a delimiting methylation-insensitive enzyme. Digested DNA was subjected to Southern hybridisation with a nptII coding region probe [indicated in (A) and (E)]. (A) The nptII region common for both constructs is depicted (refer to Fig. 1). Sites for non-methylation-sensitive limiting enzymes, NcoI for ex1 and HincII for ex2–4, are shown. (B) Methylation analysis of ex1 lines 1 and 2 after digestion with NcoI (N) or this enzyme in combination with SacII/SstII, PstI, NarI or NruI. The latter sites are methylated when the hybridising fragment has the same size as the delimiting fragment (N). The presence of this fragment and in addition smaller fragments indicate partial methylation. (C) Methylation analysis of ex2–4 lines using the delimiting enzyme HincII (H) alone or in combination with SacII. A slightly shorter hybridising band is generated when the SacII site is unmethylated. (D) Methylation analysis of ex2–4 lines using the delimiting enzyme HincII (H) alone or in combination with NarI. Note that the hybridising fragment is identical in size to the delimiting fragment for all lines. (E and F) Methylation analysis in plant:CpG-rich lines with HindIII (H) as the delimiting enzyme. The SacII (S) site was unmethylated in all lines as indicated by a hybridising fragment of 1660 bp depicted by an arrow. (G) Methylation analysis in the PvuII site in plant:CpG-rich lines using HindIII as the delimiting enzyme. Note the partial methylation in all lines as indicated by fragments that are larger than the fragment of the delimiting enzyme, marked by an arrow. Thick lines, plant DNA; N, NarI; Nr, NruI; P, PstI; Pv, PvuII; S, SacII/SstII. Asterisks, siblings of same line; TS, transgene silencing lines.