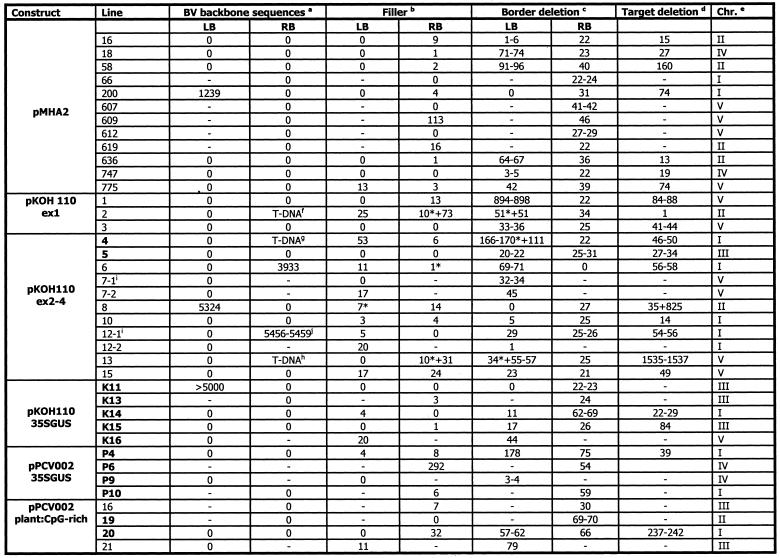
Table 2. Target and T-DNA characteristics and integration of BVB or additional T-DNA sequences of lines presented in this study.
Lines given in bold display nptII silencing. –, not determined. *Data for truncated T-DNA copies.
aNumber of base pairs upstream of the borders derived from the backbone sequence of the T-DNA vector.
bNumber of base pairs at the T-DNA/plant DNA junction that are neither a continuation of the T-DNA nor the plant-DNA.
cDeletion of the border counted from the first base in the border repeat. In cases of micro-homology between target and T-DNA, a range is given for the border and target deletion.
dNumber of base pairs deleted at the target upon integration of the T-DNA. In cases of micro-homology between target and T-DNA, a range is given for the border and target deletion.
eChromosome into which the T-DNA has integrated.
f,g,hTruncated T-DNA sequences found upstream of RB (see Fig. 2).
iCloning indicated the presence of an additional T-DNA fragment in close proximity to another T-DNA copy.
jThe BVB sequence has micro-homology at both ends.