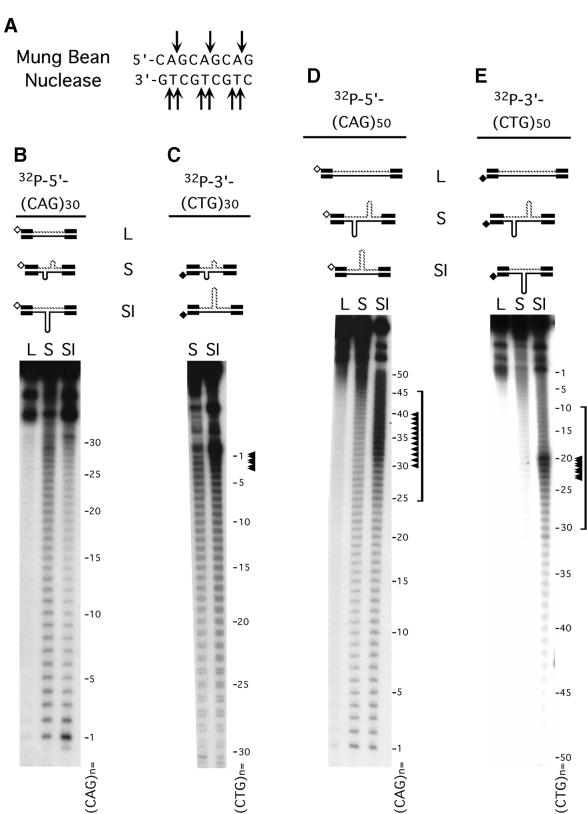
Figure 2.
S-DNAs contain regions of single-stranded DNA. (A) MBN specifically recognizes single-strand DNA; arrows indicate the observed sites of scission. (B–E) Gel-purified HindIII–EcoRI linear form (L), S-DNA (S) and SI-DNAs (SI) structures uniquely radiolabeled at the HindIII site on either the CTG or the CAG strands were digested with MBN (Materials and Methods) and products resolved on denaturing acrylamide gels. Shown are schematics of the purified DNAs and the location of the unique radiolabels, indicated by hollow or filled diamonds. Each of the complementary strands (CTG and CAG) containing either 30 or 50 repeats were individually analyzed in each of the purified structures. Sequencing ladders (not shown) indicated nucleotide positions and indicated repeat numbers are read 5′→3′ such that the first CTG repeat unit corresponds to the last CAG repeat unit. No digestion products were detected for the linear forms. Only the longer 50 repeat strands in the SI-DNAs displayed specific regions of hypersensitivity (indicated by arrowheads and brackets). While the hypersensitive sites are localized, there are more on the (CAG)50 strand than on the (CTG)50 strand. The analyses of each strand in each of the structural conformations are summarized in Figure 4.