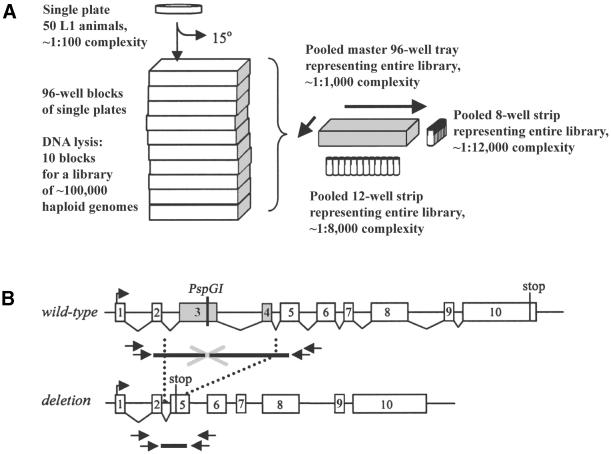
Figure 1.
Schematic of the deletion screen. (A) Method for pooling mutant worm library lysates (7,19,20). A library consisted of 960 individual plates, each seeded with approximately 50 L1 animals and allowed to grow for approximately two generations. One quarter of worms from each plate was transferred for lysis in 96-well blocks, while the remainder were maintained as dauers at 15°C. DNA lysates made from ten 96-well blocks were pooled to one master 96-well tray, increasing the genomic DNA complexity of each sample 10-fold to ∼1:1000. Lysates were further pooled into single 12-well and 8-well strips, each representing the entire library at sample complexities of 1:8000 and 1:12 000, respectively. From these two sample strips, the row and column coordinates for any positive signal were identified. (B) The PCR-screening method made use of nested primers flanking a thermostable endonuclease recognition site in the targeted gene. Wild-type product size was chosen to be ∼3 kb to allow for amplification of deletion products up to ∼2.6 kb. Digestion with thermostable restriction endonuclease allowed only deletion amplicons to be amplified. Deletions may cause a frameshift mutation, leading to premature truncation of the targeted gene, or other deleterious molecular defects. Two rounds of PCR were performed with primers designed with a high annealing temperature (∼65°C). Second round PCRs (outer primers) were performed with a dilution of the first round (inner primers) reaction.