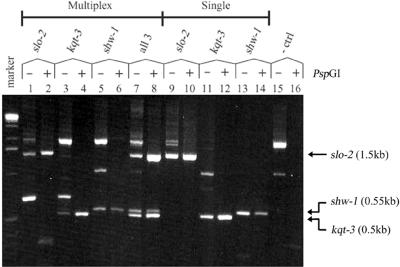
Figure 4.
Multiplexing multiple primer sets in PCR assays with PspGI digestion. PCR deletion assays were performed with three primer pairs (lanes 1–8, and 15 and 16) or single primer pairs (lanes 9–14), with and without PspGI digestion. Model templates consisted of 5 pg of genomic DNA from deletion strains slo-2(nf100) (lanes 1, 2, 9 and 10), kqt-3(aw1) (lanes 3, 4, 11 and 12) and shw-1(r1159) (lanes 5, 6, 13 and 14) added singly, or combined together (lanes 7, 8, 15 and 16), in the background of 50 ng of WT genomic DNA. PspGI digestion resolved single deletion bands corresponding to each deletion product [slo-2(nf100), 1.5 kb; shw-1(r1159), 0.55 kb; kqt-3(aw1), 0.5 kb] when all three primer pairs were multiplexed (lanes 1–8), with a specificity comparable with single primer pair controls (lanes 9–14). The presence of all three deletion templates could be unambiguously detected in single multiplexed reactions (lanes 7 and 8). PCR products resolved on a 1.2% agarose gel with a 1 kb DNA size marker (Gibco).