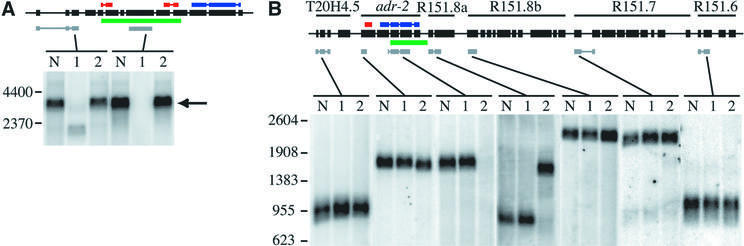
Fig. 4. Northern analyses of adr deletion strains. Black boxes with a line through the center represent the genomic structure of the adr-1 gene (A) or the six-gene adr-2 operon (B). Colored bars show the location of dsRBMs (red), catalytic domain (blue), sequences deleted in the mutants (green) and the regions of cDNA sequences used to probe northerns (gray). Gels were loaded with poly(A)+ RNA from three C.elegans strains: wild-type (lane N), adr-1(gv6) (lane 1) and adr-2(gv42) (lane 2). (A) Hybridization with a probe to the 5′ region of adr-1 shows a band corresponding to the full-length transcript (arrow) in wild-type and adr-2(gv42) samples and a shorter transcript in the adr-1(gv6) deletion strain; the strong hybridization signal of this experiment did not allow resolution of alternative splice forms. A probe to sequences within the deleted region shows equivalent expression in wild-type and adr-2(gv42) lanes and no signal in the adr-1(gv6) lane. (B) Hybridization with probes to various regions of the six-gene operon shows that transcripts from T20H4.5, R151.8b, R151.7 and R151.6 are unaffected by the deletion in adr-2(gv42); all genes are named as previously specified (Hough et al., 1999). A probe to the 5′ region of adr-2 hybridizes to a slightly shorter transcript in the adr-2(gv42) deletion strain, while a probe within the deleted region shows normal hybridization to wild-type and adr-1(gv6) RNA but no signal in the adr-2(gv42) RNA. A probe to R151.8a, the gene just downstream of adr-2, confirms that adr-2(gv42) contains a chimeric transcript that fuses adr-2 to R151.8a. The chimeric RNA is predicted to result in a chimeric protein with the first two exons of adr-2 in-frame with the protein encoded by R151.8a. An RT–PCR product spanning the junction of the adr-2::R151.8a fusion was cloned and sequenced, and confirmed results of the northern analyses.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.