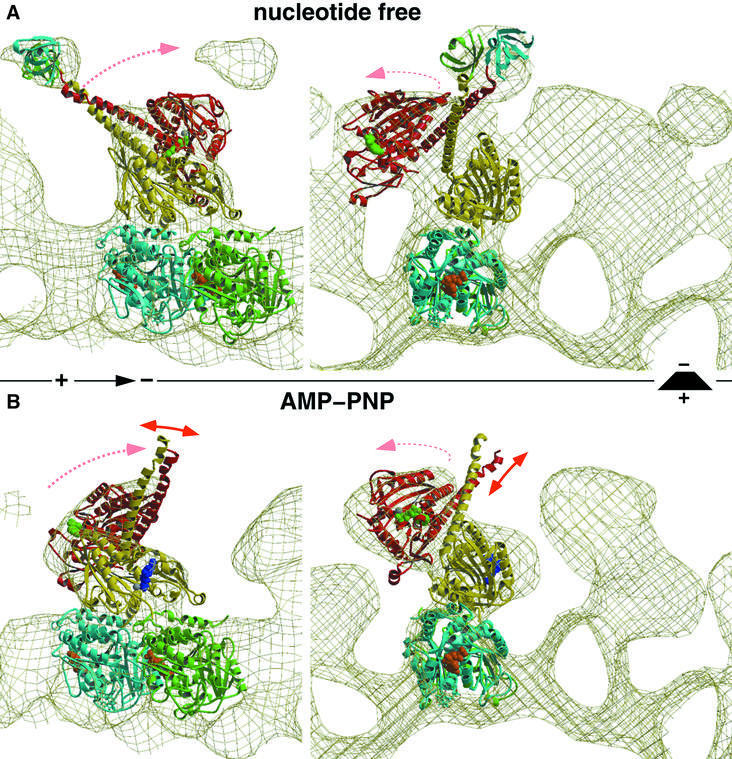
Fig. 3. Molecular docking and modeling, fitting the atomic resolution X-ray structure from Sablin et al. (1998) into our EM-derived three-dimensional maps of MT–ncd complexes. The panels on the left show side views of a protofilament with the plus end to the left. The right panels show end-on views of protofilaments viewed from the plus end. (A) The conformation of dimeric ncd in the absence of nucleotides strongly resembles the ADP state found in the crystal structure. The neck was visualized by two SH3 domains engineered to the N-terminal ends of the neck helices. (B) The neck of the untagged construct was not visible in the presence of AMP-PNP due to an increased flexibility in that region. Nevertheless, our docking experiments revealed a large conformational change, rearranging heads 1 and 2 dramatically. Head 2 rotates around a radial axis located at the C-terminal end of the neck (the transition into the core). Thereby, the two heads approach each other and close the gap towards the plus end, thus moving the neck into the minus-end direction.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.