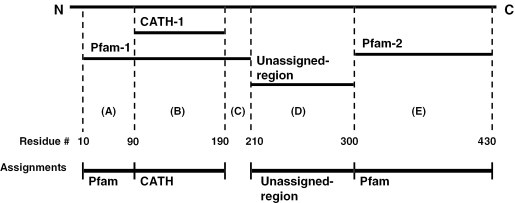
Figure 2.
Assignment of CATH, Pfam and unassigned-regions to Gene3D sequences. A hierarchical scheme is used, where CATH domains are first assigned, followed by non-overlapping Pfam domain assignments. In this example, a single CATH domain and two Pfam domains have matched a single sequence. CATH-1 and Pfam-1 overlap, with Pfam-1 extending beyond the CATH domain by 80 residues towards the N-terminus and 20 residues towards the C-terminus. According to the hierarchy, the CATH assignment (region B) is given priority and therefore assigned. Pfam-1 is then considered. When assigning domains we apply a conservative length cut-off of ≥50 residues. Consequently residues 10–89 corresponding to a fragment of Pfam-1 (region A), are assigned as a Pfam-1-subdomain whereas residues 191–201 (region C) are not. There is no such clash for Pfam-2 (region E) and this assignment is accepted. Residues 211–300 (region D) cannot be assigned to a CATH or Pfam family, yet form a sequence fragment of ≥50 consecutive residues. We refer to such sequences as ‘unassigned-regions’.