Figure 6.
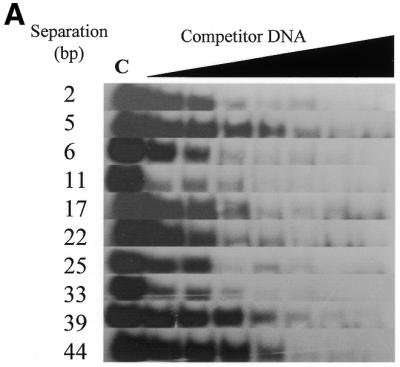
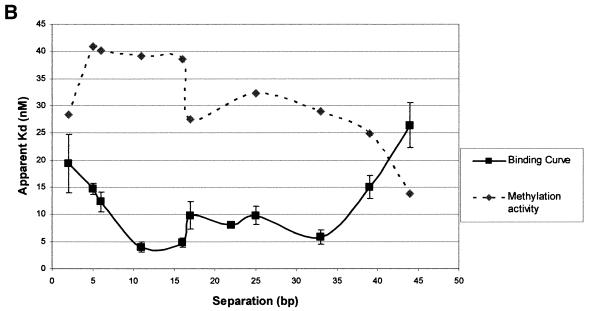
Zf.M.HpaII binds specifically to oligodeoxynucleotides with variable ‘subsite’ spacings. (A) Gel retardation analysis. Lane C in all cases contains Zf.M.HpaII enzyme and ZfHpaII labelled probe at the same levels as shown in Figure 3. Competitor DNA was added to the same final concentrations as given in Figure 3. The base pair separation between subsites for each competitor oligonucleotide is shown to the left of each gel. (B) The apparent Kd values for triplicate competition binding experiments for each subsite spacing were plotted against the subsite spacing. The methylation signal obtained from incubation of each oligonucleotide (30 pmol) with Zf.M.HpaII protein (0.1 pmol) for 30 min in the presence of [3H]AdoMet is also plotted on the same graph. The curve shown is representative of a triplicate set, which all followed the same trend. The methylation intensities have been normalised to fit on the graph.