Figure 1.
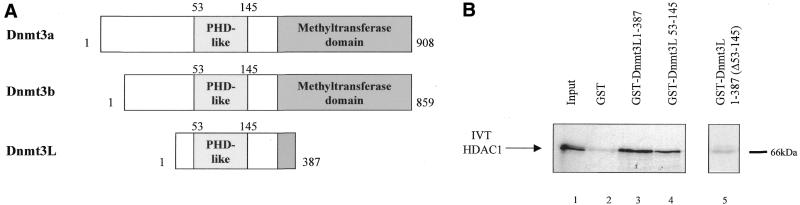
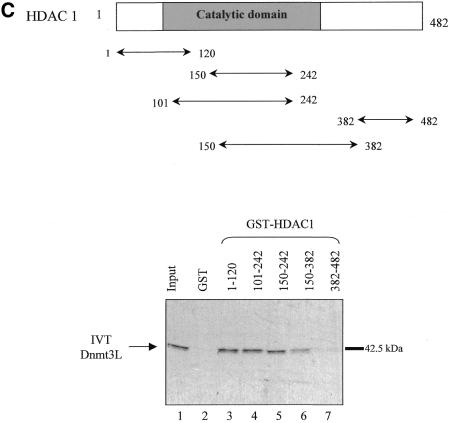
. Dnmt3L interacts with HDAC1 in vitro. (A) Schematic representation of the Dnmt3 family members. The conserved N-terminal cysteine-rich region, the PHD-like motif, characteristic of the Dnmt3 family is shown. The C-terminal domain is the catalytic methyltransferase domain present in Dnmt3a and Dnmt3b whereas the key catalytic motifs are absent in Dnmt3L. Numbers indicate amino acid residues. (B) Dnmt3L binds the histone deacetylase HDAC1 in GST pull-down experiments. Full-length HDAC1 was IVT and 35S-radiolabeled, incubated with equivalent amounts of GST (lane 2), GST fusion proteins of Dnmt3L 1–387 (full-length; lane 3), residues 53–145 (containing the PHD-like motif; lane 4) or Dnmt3L 1–387 Δ53–145 (a mutated version that lacks the PHD-like motif; lane 5). GST 1–387 Δ53–145 (lane 5) was tested in a separate experiment than the other constructs (lanes 1–4). Molecular weight in kDa is indicated on the right. The bound IVT HDAC1 is indicated by an arrow on the left. Lane 1, 35S-radiolabeled HDAC1 input (10%). (C) Dnmt3L binds specific regions of HDAC1 in vitro. The upper panel is a schematic representation of the HDAC1 deacetylase with its catalytic domain depicted by a grey box. The indicated GST–HDAC1 fusions were tested in GST pull-down experiments using IVT full-length Dnmt3L (lower panel, lanes 3–7). Lane 1, 35S-radiolabeled Dnmt3L input (10%).