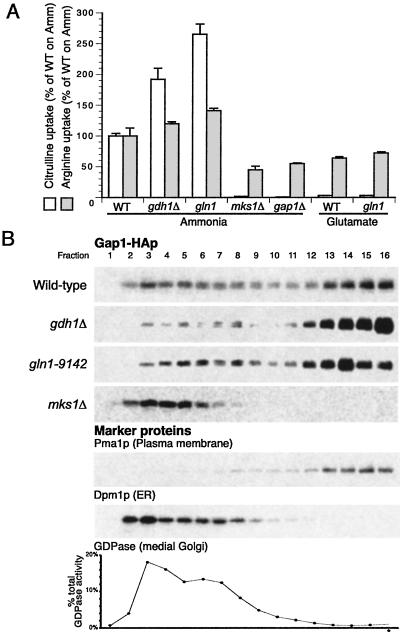
Fig 3.
Mutations that affect glutamate and glutamine synthesis pathways affect Gap1p activity and localization. (A) Gap1p activity was measured by assaying the rate of [14C]citrulline uptake in wild type (CKY759), gdh1Δ (CKY762), gln1-9142 (CKY764), mks1Δ (CKY763), and gap1Δ (CKY445), grown in ammonia or glutamate minimal medium at 24°C. All strains (except gap1Δ) contained PADH1-GAP1-HA instead of GAP1 under its own promoter. The rate of [14C]arginine uptake, which is not sensitive to nitrogen source quality, was also measured for comparison. (B) The amount of Gap1p localized to the plasma membrane is increased in gdh1Δ and gln1-9142 mutants and decreased in an mks1Δ mutant. Wild-type (CKY759), gdh1Δ (CKY762), gln1-9142 (CKY764), and mks1Δ (CKY763) cells were grown on ammonia minimal medium at 24°C, and cell extracts were subjected to isopycnic fractionation on 20–60% sucrose density gradients. Fractions were collected and proteins were separated by SDS/PAGE. Gap1-HAp was detected by immunoblotting with 16B12 anti-HA antibody. The plasma membrane marker protein Pma1p and the endoplasmic reticulum membrane protein Dpm1p were detected by immunoblotting, and the Golgi marker protein GDPase was detected by enzymatic assay. In all of the strains shown, Pma1p, Dpm1, and GDPase fractionated in the same manner. Gradients are for comparison between amounts of internal and plasma membrane-localized Gap1p in each strain and are not intended for quantitative comparison of Gap1p levels between different strains.