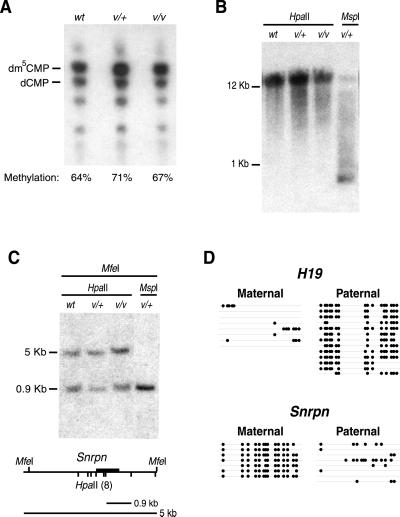
Fig 3.
Normal levels of genomic methylation in heterozygous and homozygous Dnmt1V mutant mice. (A) Similar levels of global genomic methylation among WT (wt), heterozygous Dnmt1V mutant mice (v/+), and homozygous Dnmt1V mutant mice (v/v). DNA was cleaved with the methylation-insensitive enzyme MspI before end-labeling with [γ-32]ATP, digestion to 5′-mononucleotides with nuclease P1, and separation of 5′-deoxynucleotides by TLC on cellulose plates (5). The positions of dm5CMP and dCMP are shown. The position of dCMP was determined by HpaII digestion of WT DNA (data not shown). Global genomic methylation levels are determined by the ratio of dm5CMP to total 5′-deoxycytidine (dm5CMP + dCMP). Actual experimental values are: wt = 64 ± 3%; v/+ = 71 ± 0.2%; v/v = 67 ± 3%. (B) Normal methylation of intracisternal A particle proviral sequences in genomic DNA from heterozygous and homozygous Dnmt1V mutant mice. HpaII and MspI digests of genomic DNA were hybridized with a probe to the LTR sequence of the agouti Aiapy allele (14). (C) Normal methylation of the differentially methylated domain of the imprinted Snrpn gene. Southern blot of genomic DNA digested with MfeI plus either HpaII or MspI. Restriction map of the 5′-end of the Snrpn gene is shown. Dark bar is the probe. (D) Normal methylation of the differentially methylated domains of the imprinted H19 and Snrpn genes (15, 16). H19 and Snrpn methylation patterns in a genomic DNA sample from the spleen of a heterozygous Dnmt1V/+ mouse (from a cross between a homozygous Dnmt1V female and a Mus musculus castaneus male) were analyzed by bisulfite genomic sequencing (17). Parental alleles were distinguished by single nucleotide polymorphisms between Dnmt1V mice and M. musculus castaneus mice (8, 18). Alleles are represented by horizontal lines. Positions of methylated CpG dinucleotides are indicated by filled circles. The Snrpn gene was analyzed between nucleotides 2,151 and 2,562 (GenBank accession no. AF081460) and the H19 gene was analyzed between nucleotides 1,301 and 1,732 (GenBank accession no. U19619). H19 is normally methylated at all CpG dinucleotides on the paternal allele; Snrpn is normally methylated at all CpG dinucleotides on maternal allele.