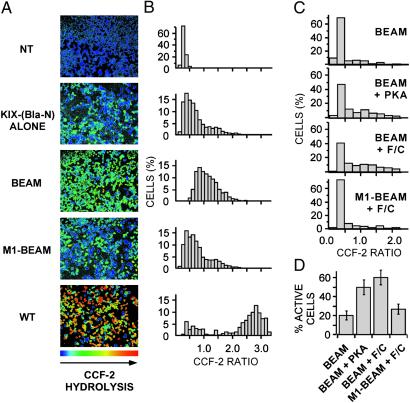
Fig 2.
293T and 293 cells expressing the KID/KIX-BEAM proteins show increased Bla enzymatic activity under conditions where the KID Ser-133 site is phosphorylated. (A) Ratiometric images of the CCF-2 fluorescence emission at 460 nm divided by the CCF-2 fluorescence emission at 530 nm (R460/530) were collected for 293T cells expressing no exogenous protein (untransfected), the KIX-(Bla-N) protein alone, the KID/KIX-BEAM proteins, the M1-KID/KIX-BEAM proteins where the KID Ser-133 site is replaced by alanine, or WT Bla. The pseudocolor images were collected after 1 h of CCF-2 loading and are shaded such that the degree of CCF-2 hydrolysis increases from blue to red (NT = untransfected; BEAM = KID/KIX-BEAM; M1-BEAM = M1-KID/KIX-BEAM; WT = WT Bla). (B) The CCF-2 R460/530 was measured for single cells in four fields (n > 1,000) for each of the conditions in A. Each bar of a histogram shows the relative percentage of cells that have a particular range of CCF-2 R460/530 values. (C) Histograms showing the percentage of 293 cells within a range of CCF-2 R460/530 values. Cells expressing either the KID/KIX-BEAM or the M1-KID/KIX-BEAM proteins were untreated (KID/KIX-BEAM; n = 123), cotransfected with constitutively active PKA plasmid (KID/KIX-BEAM + PKA; n = 332), or treated with 10 μM forskolin plus 300 μM CPT-cAMP (F/C) for 1 h (KID/KIX-BEAM + F/C, n = 254; and M1-KID/KIX-BEAM + F/C, n = 299). For cells expressing the KID/KIX-BEAM proteins, either coexpression of constitutively active PKA or treatment with 10 μM forskolin plus 300 μM CPT-cAMP caused a marked increase in the number of cells exhibiting higher levels of Bla enzymatic activity. Cells expressing the M1-KID/KIX-BEAM proteins where the KID Ser-133 is replaced by alanine were indistinguishable after stimulation with 10 μM forskolin and 300 μM CPT-cAMP (M1-KID/KIX-BEAM + F/C) from untreated cells expressing the KID/KIX-BEAM proteins (KID/KIX-BEAM). (D) The percentage of cells exhibiting Bla enzymatic activity for each of the conditions presented in C. Cells were judged to be enzymatically active if the average cellular R460/530 was >0.4, which corresponded to the average R460/530 observed in untransfected cells.