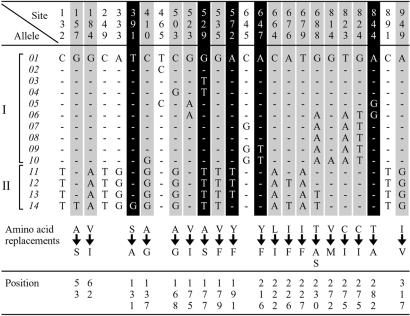
Fig 3.
Variable nucleotide sites differentiating LWS alleles of Lake Victoria/Lake Nabugabo cichlids. A dash (—) indicates identity with the top sequence (allele 01). Nonsynonymous substitution sites in which amino acid replacement might change the absorption spectra of opsin proteins are highlighted in black; the remaining nonsynonymous sites are highlighted in gray. At 3 of the 19 nonsynonymous sites (391, 529, and 844) the amino acid replacements caused by the nucleotide substitutions have been shown in other systems to result in shifts of absorption sensitivity (λmax; refs. 26 and 27). At one of the three sites (391) and two other sites (572 and 647) substitutions cause amino acid replacements at positions 131, 191, and 216, respectively, which correspond to positions 118, 178, and 203 in the bovine rhodopsin molecule, respectively. From the crystal structure of bovine rhodopsin (28), the residues at positions 131, 191, and 216 in the cichlid LWS opsin can be expected to be close to each other and may be able to form hydrogen bonds among them. The replacements, in cichlid LWS, could be expected to disrupt these hydrogen bonds and thus lead to a shift in absorption sensitivity. Substitutions at sites 529 and 844 cause amino acid replacements at positions 177 and 282, respectively, which are among the five positions that, according to the “five-sites rule” of Yokoyama and Radlwimmer (27), are primarily responsible for the spectral sensitivities of the MWS and LWS pigments in humans and fishes. The two ancient allelic lineages (I and II) are indicated by brackets. Amino acid replacements are given in the International Union of Physiological Sciences single-letter code; the arrows indicate a change from the more common to the less common residue. “Positions” refers to the numbering of amino acid residues in the cichlid LWS opsin.