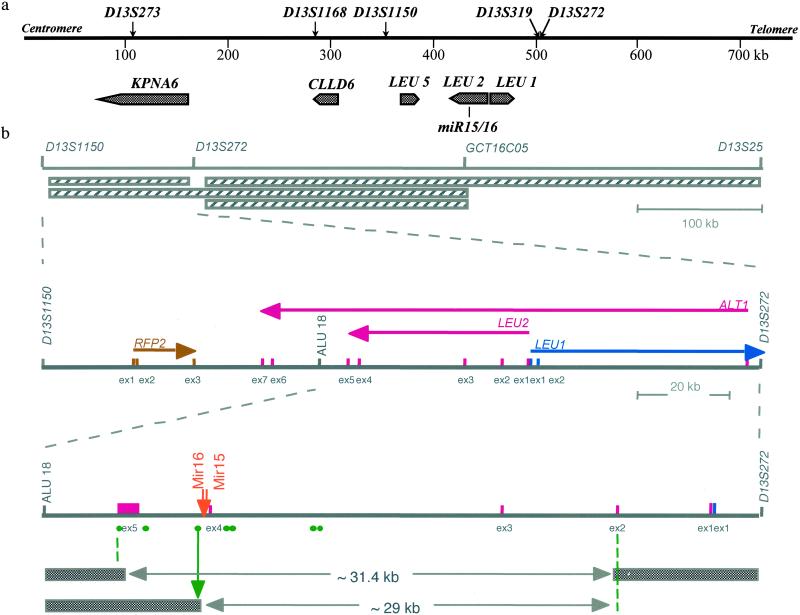
Fig 1.
(a) Genes within the 13q14 tumor suppressor locus in CLL and localization of miR15/16 cluster. The positions of genetic markers and the positions of genes on the map are shown. (b Top) Map of the locus with previously reported 13q14 deletions marked by horizontally striped boxes. (Middle) Map of the locus between D13S1150 and D13S272 markers. The orientation of each gene is marked by an arrow under the gene's name. Colored vertical bars mark the position of corresponding exons for each gene. (Bottom) Map of the locus between Alu18 and D13S272 markers. Bars and boxes mark the position of exons for LEU2/ALT1 and LEU1 following colors in panels above. The orange arrow marks the position of miR15 and miR16 genes. Green circles mark the positions of PCR primers used to screen somatic cell hybrid clones derived from a fusion of two independent leukemia cases (CLL-A and CLL-B). Green circles mark the positions of oligonucleotides pairs used to screen hybrids by PCR. All oligonucleotides pairs shown, as well as exon-specific primers for each exon shown, and miR15/16-specific primers were used to screen the hybrids. The green arrow represents the position of the breakpoint in CLL-B carrying a t(2;13)(q32;q14) translocation. Filled boxes represent portions of chromosome 13 present in the hybrids. The 31.4-kb deletion was present in a clone derived from CLL-A, a patient with CLL carrying a t(2;13)(q12;q13) translocation, bilateral retinoblastoma, and ulcerative colitis.