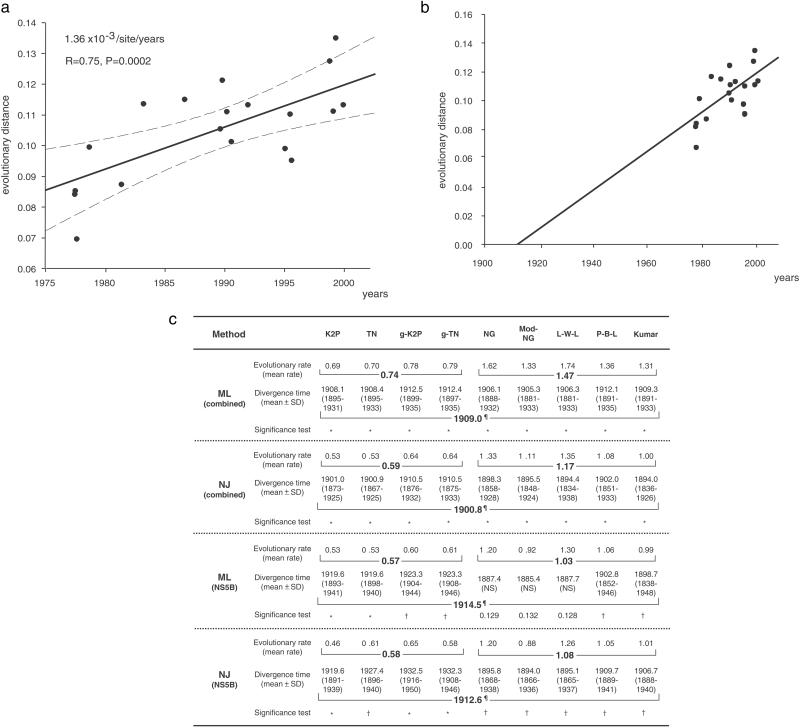
Fig. 2.
Estimating the most recent common ancestor of genotype 1a in the U.S. based on data collected over the last three decades. (a) A regression analysis within combined regions was performed to estimate a mean molecular clock. The mean evolutionary rates (solid regression line) of the P-B-L model indicated 1.36 × 10−3 per site per year (P = 0.0002). The 95% confidence intervals of the regression line are indicated by broken lines. (b) The divergence time of the most recent common ancestor of U.S. genotype 1a was estimated to have occurred around 1910, which is supported by most evolutionary models (c). (c) By using the mean molecular clock derived from regression analyses of serially determined phylogenetic trees, the divergence time of the most recent common ancestor of genotype 1a in the U.S. was estimated. Models for evolutionary analyses are explained in Methods. Phylogenetic trees were analyzed by the maximum likelihood and the NJ methods. To estimate the divergence time (mean ± SD), 500 bootstrap replicates were generated by random-with-replacement resampling of the data points to determine the molecular origin of ancestral sequences. Mean and SD of the divergence times also were determined. Standard significance testing was conducted. *, P < 0.01; †, P < 0.05; ¶, mean of 95% significance divergence time (year); NS, not significant. Evolutionary rate = number of nucleotide substitutions × 10−3 per site per year.