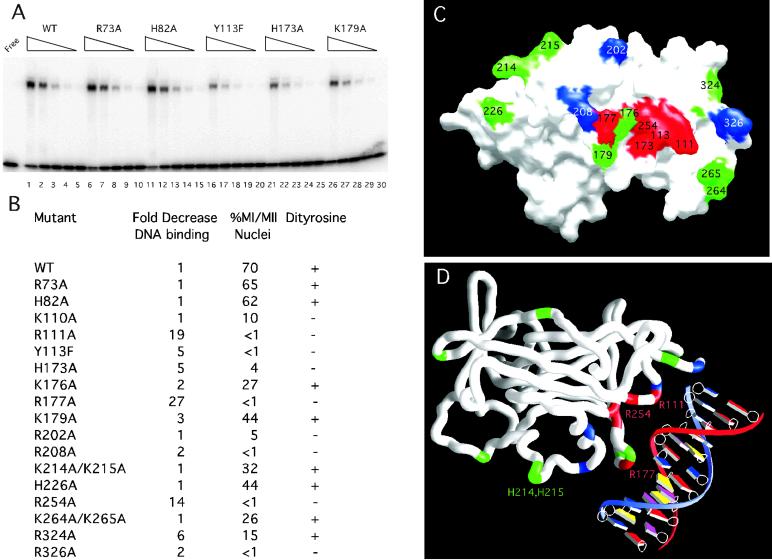
Fig 3.
Mutational analysis and modeling of Ndt80–DNA interactions. (A) The DNA-binding activity is shown for the WT and mutant Ndt80 proteins. WT (lanes 1–5), R73A (lanes 6–10), H82A (lanes 11–15), Y113F (lanes 16–20), H173A (lanes 21–25), and K179A (lanes 26–30) Ndt80 proteins were purified and assayed by EMSA. Lanes 1, 6, 11, 16, 21, and 26 contain the same amount of purified protein. The wedges indicate decreasing concentrations of each protein corresponding to fivefold serial dilutions in successive lanes. (B) The relative decrease in DNA-binding activity analyzed by EMSA, progression through meiosis I and II (MI/MII), and spore-wall formation (Dityrosine) was compared for 18 mutant Ndt80 proteins and the WT protein. (C) A molecular surface rendering of the DNA-binding domain of Ndt80 is shown looking directly at the proposed DNA-interacting surface of the molecule. Residues shown in red result in at least a fivefold decrease in relative DNA-binding affinity and loss of the ability to sporulate in vivo when substituted with Ala or Phe in the case of Y113. Those in green have near WT DNA-binding activity and retain the ability to sporulate, and those shown in blue retain near WT DNA-binding activity but fail to sporulate in vivo. Note: 12 residues in the A molecule for the form II structure lack clear side chain density (including 176, 177, and 179) and have been modeled as Ala. (D) A model for Ndt80 bound to the MSE reveals a previously uncharacterized DNA-binding motif. A worm rendering of the protein is shown with the DNA. Residues R111, Y113, H173, R177, and R254 (red) are predicted to interact with the DNA based on the model and the mutational analysis. Residues in green are not predicted to interact significantly with the DNA. Those in blue may have a role distinct from DNA binding that is required for activation in vivo, i.e., they may interact with other proteins.