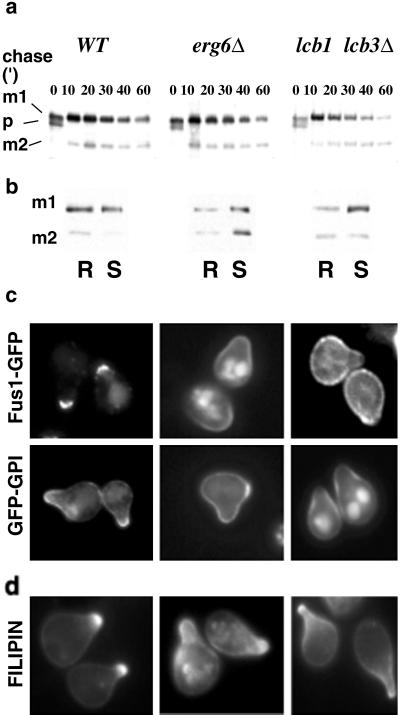
Fig 5.
Processing and sorting of shmoo-tip markers in lipid biosynthetic mutants. (a) WT (MBY206), erg6Δ (MBY240), and lcb1–100 lcb3Δ (MBY223) cells were preincubated with 5 μM α-factor for 15 min, pulse-labeled with [35S]methionine for 5 min, and chased for various times at 24°C. Fus1-PA maturation was analyzed by immunoprecipitation, SDS/PAGE, and autoradiography. The position of precursor (p) and to mature products (m1 and m2) is indicated. (b) DRM association of Fus1-PA in WT (MBY206), erg6Δ (MBY240), and lcb1–100 lcb3Δ (MBY223) cells was analyzed as in Fig. 2. (c) Localization of Fus1-GFP (pMBQ30) and GFP-GPI (pMBQ31) in WT (RH690-15D), erg6Δ (RH3622), and lcb1–100 lcb3Δ (RH690-13B) cells. Cells were grown on YPGal and treated with 3 μM α-factor for 3 h. (d) Sterol distribution.