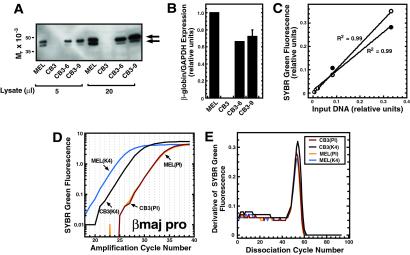
Fig 2.
Quantitative ChIP analysis reveals differential H3-meK4 at the βmajor promoter in MEL and CB3 cells. Cells were incubated for 4 days with 1.5% DMSO. (A) p45/NF-E2 expression in DMSO-induced CB3, MEL, CB3-6, and CB3-9 cells. The arrows depict p45/NF-E2 isoforms. (B) Quantitative real-time RT-PCR analysis of β-globin expression in CB3, MEL, CB3-6, and CB3-9 cells. Relative levels of β-globin expression were normalized to the expression of GAPDH. The graph depicts data from three (MEL, CB3, and CB3-9) and two (CB3-6) independent experiments. (C) SYBR green fluorescence (relative units) was plotted vs. the initial MEL and CB3 cell input DNA concentration. The plot illustrates the linearity and range of signals used to measure the relative amounts of target DNA in samples. (○) CB3 cells. (•) MEL cells. (D) A representative amplification plot for quantitative analysis of H3-meK4 at the βmajor promoter in MEL and CB3 cells. PI, preimmune. (E) Dissociation curves of the amplicons illustrated in D. The homogeneity of the curves reflects the generation of a single amplicon.