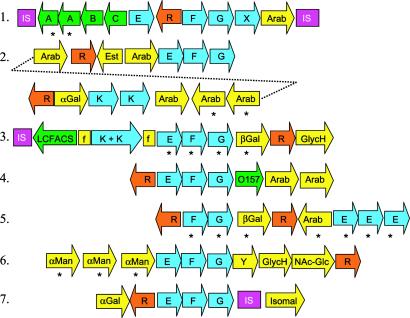
Fig 2.
Oligosaccharide utilization gene clusters. Genes are represented by arrows. IS, insertion sequence; F and G, MalF-type and MalG-type permease subunits of ABC transporter, respectively; E and K, MalE-type solute binding protein and MalK-type ATP-binding protein of ABC transporter, respectively; R, LacI-type repressor; Arab, arabinosidase; βGal, β-galactosidase; αMan, α-mannosidase; αGal, α-galactosidase; GlycH, glycosyl hydrolase of unknown specificity; Isomal, isomaltase; NAc-Glc, N-acetyl glucosamindase; O157, ORF with homolog only in E. coli O157; X and Y, unique hypothetical proteins; LCFACS, long chain fatty acyl CoA synthetase; Est, possible xylan esterase; A, L. lactis phage infection protein homolog; B, oxidoreductase; C, phosphoglycerate mutase; f, fragment of AraE permease. Asterisks mark recent gene duplications.