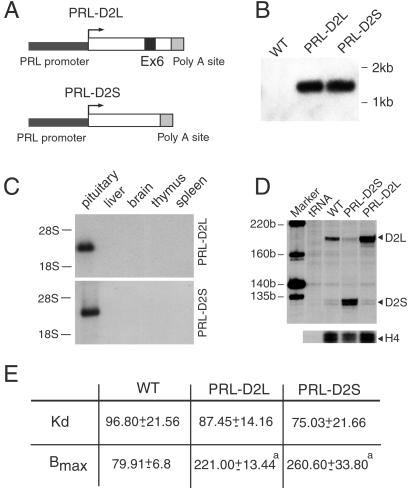
Fig 1.
Analyses of PRL-D2L and PRL-D2S transgenic mice. (A) Schematic representation of the chimeric constructs used to generate the PRL-D2L and PRL-D2S transgenic lines. Exon 6 (black box) is absent in the D2S cDNA. The 3-kb promoter region of the rat PRL gene is indicated in black. The polyadenylylation site of the SV40 large T antigen (gray box) has been added at the 3′ of each construct. (B) Southern blot analysis of genomic DNA from tail biopsies from WT, PRL-D2L, and PRL-D2S mice. Genomic DNA was digested by PstI, blotted, and hybridized by using the SV40 poly(A) sequence as probe. (C) Two micrograms of total RNA from the pituitary gland and 10 μg of all of the other tissues of animals of each genotype were used for Northern blot analysis. The filter was hybridized with a 32P-labeled probe corresponding to the SV40 poly(A) sequence. (D) RNase protections were performed by using 5 μg of total RNA hybridized with a mouse D2R specific 32P-labeled probe. The size of protected fragments was 202 bp (D2L) and 131 bp (D2S) as indicated. A histone H4 probe was used as an internal control. (E) [3H]Spiperone binding on membrane extracts from pituitaries of WT, PRL-D2L, and PRL-D2S mice. Nonspecific binding was determined in the presence of (+)-butaclamol (1 μM). The affinity (Kd) and the maximal number of binding sites (Bmax) were calculated on Scatchard's transformation of saturation curves. The Kd is expressed in picomolar and the Bmax in femtomoles of D2 receptor per mg protein. Values are mean ± SEM of three different experiments performed in duplicates. a = P < 0.001 (***) versus wild-type mice (Student's t test).