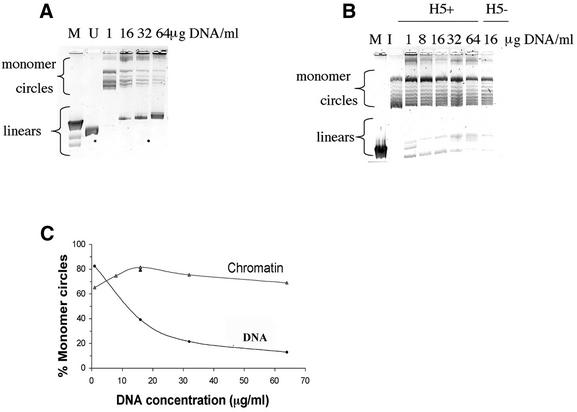
Figure 2.
DNA products obtained from a linear 2.7 kb pUC19 DNA fragment treated with T4 DNA ligase in the form of naked DNA or chromatin at increasing DNA concentrations. The 2% agarose gels contained 0.2 µg/ml chloroquine sulfate in order to resolve the topoisomers resulting from the closed circular DNA forms. Monomer circle and linear DNA forms are indicated. (A) Lanes labeled 1–64 µg DNA/ml show the ligation products resulting from the naked DNA. M denotes linear λHindIII DNA size markers; the 2.0 and 2.3 kb bands are resolved, whereas the 6.6, 9.4 and 23 kb bands run together. The lane labeled U denotes the unligated 2.7 kb DNA. (B) Lanes labeled 1–64 µg DNA/ml show the DNA products obtained from the H5+ chromatin. One chromatin sample lacking histone H5 (H5–) ligated at 16 µg DNA/ml is shown. The unligated DNA starting material used in this experiment is shown in A, lane U. Additionally, a core histone reconstituted DNA sample at a concentration of 16 µg/ml that was incubated overnight under ligation conditions, but without the T4 DNA ligase, ran indistinguishably from the starting DNA (not shown). M denotes linear unresolved 4.4–23 kb λHindIII DNA markers; I denotes the (Form I) supercoiled form of pUC19 having one supercoil per ∼200 bp. (C) Quantitation of the percentage monomer circles formed for DNA or chromatin ligated over a range of DNA concentrations. The single black triangle at 16 µg/ml denotes the sample lacking histone H5.