Figure 8.
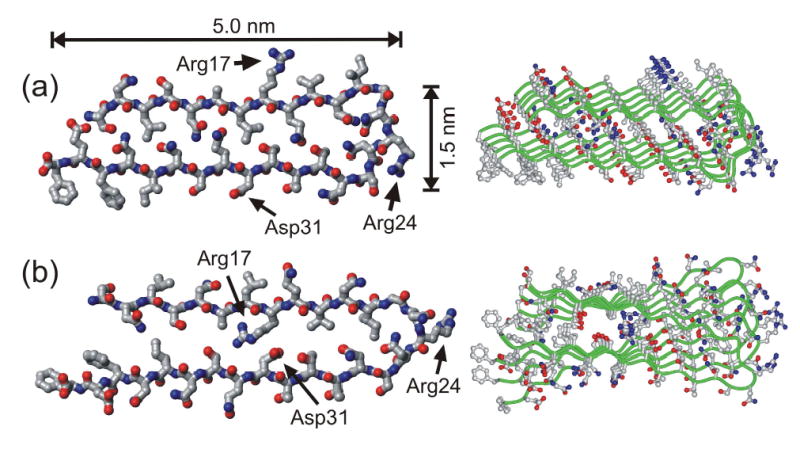
Structural models for Ure2p10–39 fibrils generated by restrained molecular dynamics and energy minimization simulations for a pentameric assembly of Ure2p10–39 molecules. Both the final energy-minimized structure of the pentamer (right) and the conformation of the central molecule in the pentamer (left) are shown. The two models differ in the relative orientation of the β-sheets formed by the two β-strand segments (residues 10–21 and 27–39), as dictated by the conformation of the intervening loop segment (residues 22–26). (a) Model in which all charged sidechains are outside the fibril core. (b) Model in which oppositely charged sidechains of Arg17 and Asp31 can form internal salt bridges in the fibril core.
