Fig. 1. Assays measuring supercoil domain structure in Salmonella and E. coli.
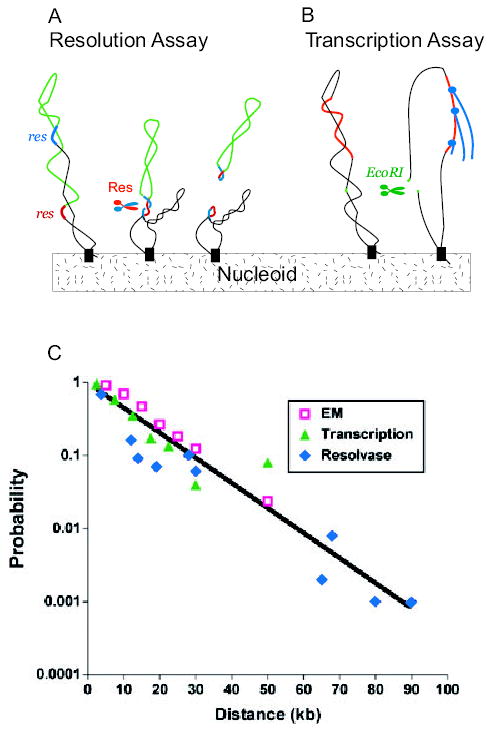
A. A set of Salmonella strains were made with a pair of directly repeated 140 bp Tn3 res sites (red and blue DNA) separated by different segments of bacterial DNA (green). After expressing the γδ site-specific recombinases (Res), a deletion forms if and only if the res sites reside in a common supercoil domain (Stein et al., 2005).
B. Escherichia coli cells contain 300 supercoil responsive genes (SSGs) whose expression is rapidly increased or decreased by a loss of negative supercoiling (Peter et al., 2004). Expression of EcoRI causes double-strand breaks and loss of supercoiling from domains with a restriction site. Changes in RNA levels measured with whole genome microarrays measure supercoil diffusion when data are combined with distance from the promoter to restriction sites in the genome (Postow et al., 2004).
C. The probability of supercoil loop detection as a function of DNA length in kb is plotted for resolution assays (blue diamonds) SSG transcription data (green triangles) and loop sizes measured from EM images (red squares). (Lisa Postow provided the transcription data and plot.)
