Figure 5. ATHB15 mRNA cleavage by miR166 in Nicotiana benthamiana, wheat germ extract, and in Arabidopsis.
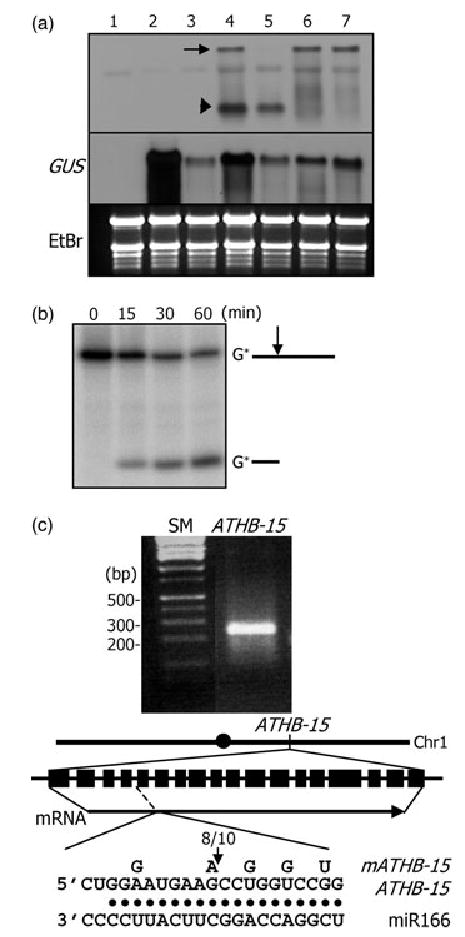
(a) ATHB15 mRNA cleavage in N. benthamiana. Total RNAs were extracted from plants without any vector constructs (1), with pBI121 control alone (2), with pBI121 + miR166 vector (3), with pBI121 + ATHB15 vector (4), with pBI121 + ATHB15 vector + miR166 vector (5), with pBI121 + mATHB15 vector (6), and with pBI121 + mATHB15 vector + miR166 vector (7). RNA gel blots were probed with the 5′-end cleavage fragment of ATHB15. The arrow marks the full-size mRNA, and the arrowhead the 5′-end cleavage product.
(b) ATHB15 mRNA cleavage in wheat germ extract. A 5′-end labeled ATHB15- specific RNA was used in the assay.
(c) 5′-RACE to determine the cleavage site in Arabidopsis. The arrow in the diagram marks the cleavage site. The number refers to that of independent clones with the 5′-end as determined.
