Fig. 3.
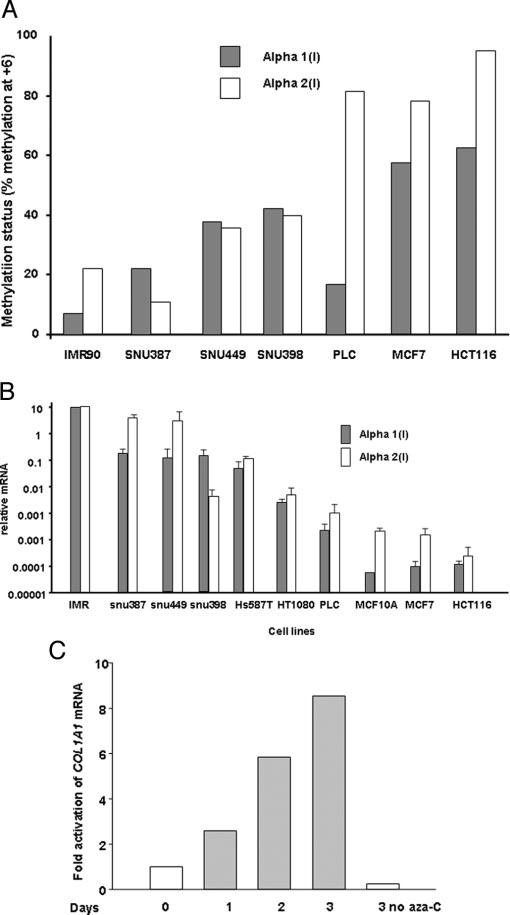
Methylation status at the RFX1 site in human cancer cells is inversely proportional to steady state mRNA level of the COL1A1 and COL1A2 genes. A, methylation status at the RFX1 site in human cancer cells. The methylation status at +7 of COL1A1 and COL1A2 genes was compared using normal (IMR90) and different cancer cell lines. Genomic DNA from the cell lines were bisulfite modified, amplified using converted primers, and the product was gel purified using 2% agarose gel. The isolated product was then subjected to primer extension (methylation sensitive-single nucleotide primer extension) using single radioactive dNTP and run in 7 m urea, 15% acrylamide sequencing gel at room temperature. The gel was partially dried and autoradiographed for 1 h at −80 °C. The percent methylation was determined as described under “Materials and Methods.” In the majority of cases both COL1A1 and COL1A2 genes are coordinately methylated in cancer cell lines compared with normal cell lines. B, steady state mRNA level of COL1A1 and COL1A2 genes in human cancer cells. Methylation status of the collagen gene at the +7 site correlated with mRNA expression. Steady state levels of COL1A1 and COL1A2 mRNA were measured using real-time RT-PCR. Primers used for these experiments are found in Table III. Relative mRNA for each cell line (bars) was normalized for 18 S rRNA and calculated using the 2[-Delta Delta C(T)] method (21, 68) comparing mRNA amounts measured from each cell line to mRNA amounts from normal unmethylated fibroblast (IMR-90). C, the aza-dC can reactivate the COL1A1 gene. A time study indicated the reactivation of COL1A1 gene in HT1080 cells after treatment of 1 μm aza-dC. Cells were treated with 1 μm aza-dC every day for 3 days, total RNA was isolated and reverse transcribed. Real-time RT-PCR was performed with collagen-specific primers and Taqman probe as presented in Table III. Reactivation of the COL1A1 gene was observed during 3 days of aza-dC treatment.
