Fig. 1. Alignment of all mouse CLCA isoforms.
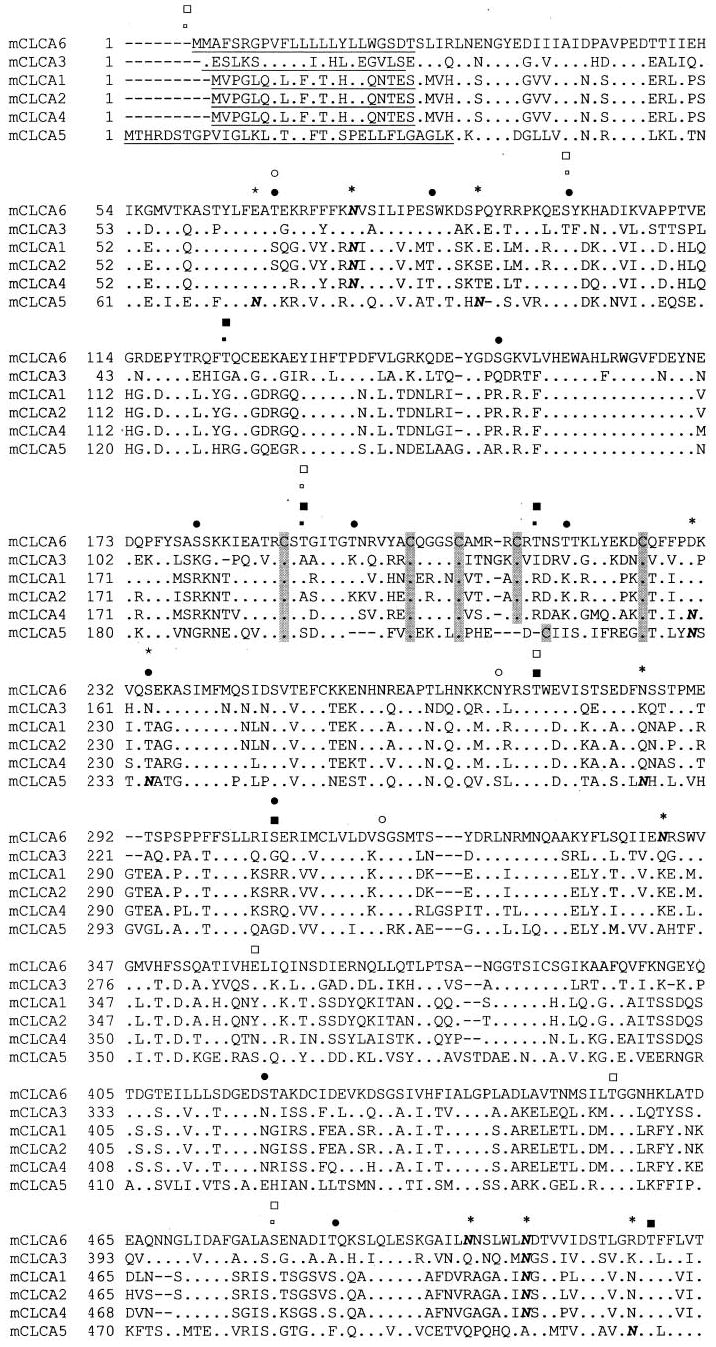

Consensus sites for phosphorylation by Ca2+/calmodulin-dependent protein kinase II, PKA, and PKC are marked for mCLCA5 and mCLCA6 (small open square, mCLCA5 Ca2+/calmodulin-dependent protein kinase II; large open square, PKA; open circle, PKC; small filled square, mCLCA6 Ca2+/calmodulin-dependent protein kinase II; large filled square, PKA; filled circle, PKC (PhosphoBase version 2.0, Center for Biological Sequence Analysis). Sites are marked for all mCLCAs for monobasic proteolytic cleavage (↓), signal sequence (underlined) (SignalPV1.1, Center for Biological Sequence Analysis), and multiple cysteine motif (C). Additionally, consensus sites for N-linked glycosylation are indicated (N with * overhead). The alignment was generated by the Clustal method.
