Figure 3.
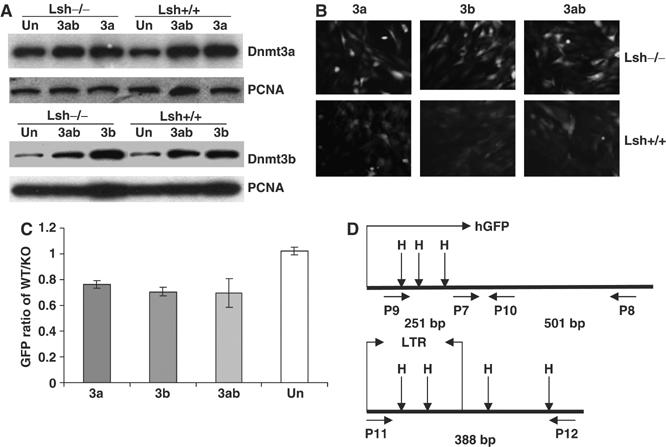
Lsh is required for silencing of Dnmt3a or Dnmt3b mediated silencing of a retroviral transgene. (A) Western blot analysis using nuclear extracts derived from Lsh−/− and Lsh+/+ mouse embryonal fibroblasts stably expressing Dnmt3a, Dnmt3b, Dnmt3a/Dnmt3b, or untransfected (Un) MEFs. For detection, specific antibodies were used against Dnmt3a, Dnmt3b, or PCNA as control. (B) Fluorescence analysis. Lsh+/+ and Lsh−/− MEFs that were stably expressing Dnmt3a, Dnmt3b, or Dnmt3a/Dnmt3b were infected with pMSCV-hGFP and examined after 8 days for GFP expression using a fluorescence microscope. (C) FACS analysis. The GFP intensity of Lsh+/+ and Lsh−/− MEF cells expressing Dnmt3a, Dnmt3b, Dnmt3a/Dnmt3b, or untransfected (Un) was measured by FACS analysis 8 days after retroviral infection. The difference in fluorescence intensity was expressed as GFP ratio of Lsh+/+ over Lsh−/−. (D) Map of the retroviral vector pMSCV-hGFP indicating the location of HpaII/MspI sites and the position of the primers used for methylation-sensitive PCR analysis. Primer pair P9/P10 detects methylation within the GFP region. Primer pair P7/P8 serves as internal control. Primer pair P11/P12 detects methylation in the 5′-LTR and the adjacent region. The length of the expected PCR fragments is indicated in base pairs (bp).
