Figure 6.
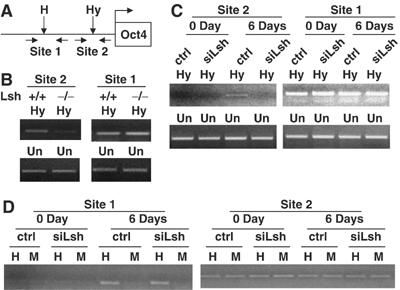
Lsh is involved in de novo methylation at the Oct-4 gene. (A) Map of murine Oct4 gene and its promoter indicating the location of HpaII/MspI site (H in Site 1) and HpyCh4 IV (Hy in Site 2) and the position of the primers used for methylation-sensitive PCR analysis. Due to different recognition sequences of HpaII and HpyCh4 IV, site 1 and site 2 can be used as internal control for each other in methylation-sensitive PCR. (B) Methylation-sensitive PCR. Genomic DNA derived from the Lsh wild type and Lsh−/− MEF cells was digested with HpyCh4 IV and subjected to PCR analysis with the indicated primer pairs. Site 1 served as internal control for site 2. Undigested DNA (Un) was used for adjustment of DNA input before digestion. (C) Methylation-sensitive PCR. Genomic DNA derived from the siLsh and control ES cells at 0 day and 6 days after differentiation was digested with HpaCh4 IV and subjected to PCR analysis with the indicated primer pairs. Site 1 served as internal control for site 2. Undigested DNA (Un) was used for adjustment of DNA input before digestion. (D) Methylation-sensitive PCR. Genomic DNA derived from the siLsh and control ES cells at 0 day and 6 days after differentiation was digested with HpaII and MspI and subjected to PCR analysis with the indicated primer pairs. Site 2 served as internal control for site 1.
