Figure 4.
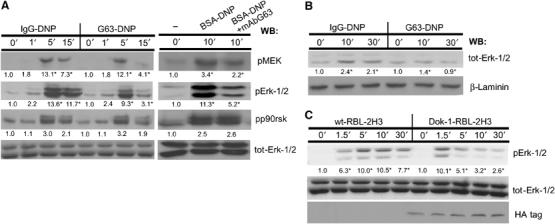
MAFA (co)-clustering interferes with the FcɛRI activation of the Erk-1/2 coupling pathway. (A) RBL-2H3 cells were treated for the indicated times with either IgG-DNP3 or G63-DNP3 (left panel). In an additional set of experiments, the cells were either FcɛRI stimulated (BSA-DNP) or first pretreated with 50 nM mAb G63 F(ab′)2 for 5 min and then FcɛRI stimulated (BSA-DNP+G63) (right panel). Cells were lysed and lysates' samples containing equal protein amounts were analyzed by WB using antibodies specific to the active forms of the following proteins: pMEK, pErk-1/2, pp90rsk, and total Erk-1/2. (B) RBL-2H3 cells (1 × 107) were treated for the indicated times with either IgG-DNP3 or G63-DNP3. Cells were lysed with Triton-based buffer and the cells' nuclei were separated and then resuspended with nuclear extract buffer. The nuclear supernatants were then separated by SDS–PAGE and WB was carried out as described above using antibodies specific to tot-Erk-1/2 and laminin. (C) Parental RBL-2H3 cells (wt-RBL-2H3) or their mutants overexpressing Dok-1 (Dok-1-RBL-2H3) were treated for the indicated times with 1 nM G63-DNP3 and lysed. Equal amounts of protein samples were taken from cell lysates, separated by SDS–PAGE, and electrotransferred onto nitrocellulose membranes. Erk-1/2 activation and the levels of proteins were monitored by sequential WB with antibodies specific to phosphorylated Erk-1/2 (pErk-1/2), total Erk-1/2 (Erk-1/2), or the HA tag (HA). Detection was carried out by ECL and quantification of fold change (in AU) was by densitometric analysis. The results represent one set out of three independent experiments; numbers highlighted with an ‘*' indicate a change that was reproduced three times (A–C).
