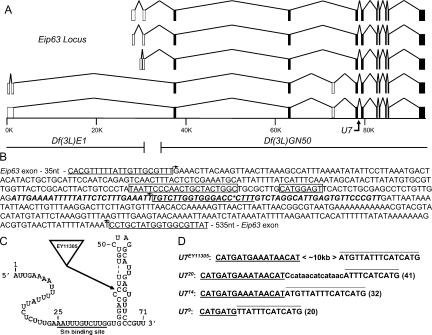
FIGURE 1.
Mutation of the Drosophila U7 gene. (A) Schematic of the Eip63 locus. The five identified Eip63 mRNAs species are shown, with open boxes indicating variably spliced exons and closed boxes indicating invariant exons. The scale bar indicates genomic DNA in kilobase pairs. The U7 gene is located at position ~79K. The right breakpoint of Df(3L)E1 and the left breakpoint of Df(3L)GN50, located at position ~31K and ~34K, respectively, are shown below the scale bar. (B) Sequence of the genomic fragment used to rescue the U7 mutant phenotype. The U7 snRNA transcript is indicated in bold italic. The asterisk indicates the location of the ~10-kb EY11305 P element. The large and the small boxes, respectively, indicate the PSEA and PSEB essential snRNA promoter elements (Zamrod et al. 1993). The bent arrows indicate primers used to analyze the excision mutations. Note that the middle and top primers were able to detect a product in U723 homozygous mutant animals. (C) Putative U7 snRNA secondary structure indicating the location of the EY11305 insertion. The underlined sequence indicates the Sm protein binding site. (D) Molecular characterization of U7 imprecise excision mutations. Each sequence shown (total length in parentheses) is located at the EY11305 insertion site indicated in panels B and C. The underlined and overlined sequence is derived from the 5′ and 3′ end, respectively, of the P element inverted terminal repeat (ITR). The lowercase sequence in U720 is not derived from the ITR, and its origin is unknown.