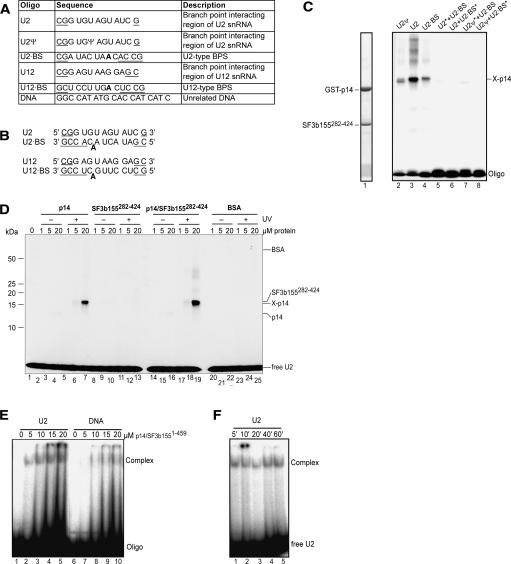
FIGURE 4.
RNA cross-linking and gel-shift assays of p14 and p14–SF3b155. (A) Sequence and description of RNA and DNA oligonucleotides. Additional nucleotides added to stabilize duplex formation are underlined. The branch point adenosine is shown in bold. (B) Duplexes formed by oligonucleotides shown in A. (C) UV cross-linking of p14–SF3b155282–424. RNP complexes were formed on the oligonucleotides or their duplexes (as indicated above the lanes). Following UV-irradiation, samples were separated on 12% SDS-PAGE and stained with Coomassie blue (lane 1) or visualized by autoradiography (lanes 2–8). The position of noncross-linked (input) proteins is indicated on the left and that of oligonucleotides and the RNA–p14 cross-link are indicated on the right. Duplexes contained only one 32P-labeled oligonucleotide, which is marked by an asterisk (*). Note that in this experiment, the p14 protein contained a GST tag. (D) UV-cross-linking of p14, SF3b155282–424, or p14–SF3b155282–424 to the U2 oligonucleotide. Increasing concentrations of protein (as indicated above each lane) were incubated with 32P-labeled U2 oligonucleotide and were either analyzed directly without UV irradiation (lanes 2–4, 8–10, 14–16, 20–22) or subjected to UV irradiation (lanes 5–7, 11–13, 17–19, 23–25) and analyzed as in C. BSA (lanes 20–25) was used as a control. (E) EMSA of the p14–SF3b1551–459 complex with oligonucleotides. Increasing concentrations of the p14–SF3b1551–459 protein complex (as indicated above each lane) were incubated with 32P-labeled U2 snRNA oligo (lanes 1–5) or unrelated DNA oligonucleotide (DNA) (lanes 6–10), separated on a native gel, and visualized by autoradiography. (F) Time course of U2 oligo-p14–SF3b1–459 complex formation. p14–SF3b1551–459 was incubated with 32P-labeled U2 oligo for the indicated time and RNP complex formation was analyzed as in E.