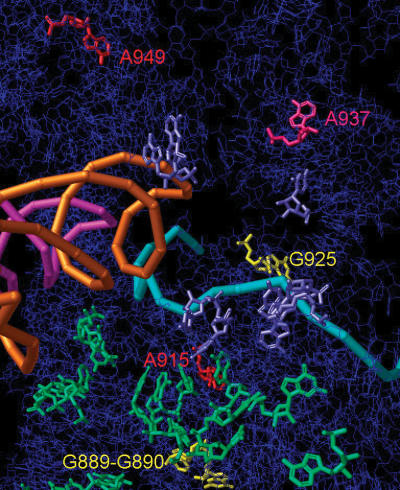
Figure 5.
Mapping of protected bases. RbbA chemical protected footprinted bases were mapped on the public data base 1YL4 structure, which is made up of Thermus thermophilus bases but with E.coli numbering (20). The 16S rRNA backbone is shown in dark blue. The 16S rRNA bases that contact the 50S subunit are enlarged and in light green. Nucleotide bases that contact the E-site RNA are shown as enlarged purple bases. Enhanced truncated RbbA protected bases A949 and A915 are in red and A937 is in pink. Base A937 was also a base that has direct contact with the E-site tRNA (20). The pink and orange spirals represent A-site and P-site tRNAs, respectively. The cyan atomic structure is the mRNA that runs though the 30S subunit. Previous RbbA protected bases G889, G890 and G925 are shown in yellow. If a structure were available for the E-site tRNA, it would be right of the orange spiral tRNA and contacting the enlarged purple bases as well as A937.