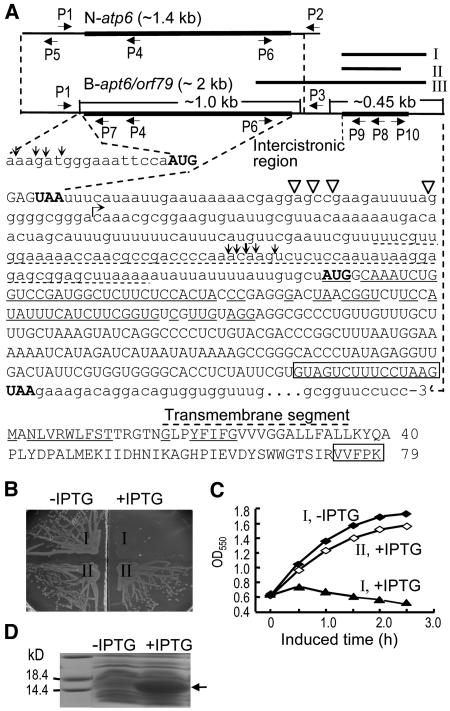
Figure 1.
Characterization of the CMS-Associated Gene in Rice with BT-Cytoplasm.
(A) The structures of N-atp6 and B-atp6/orf79 transcripts, the sequence downstream of B-atp6, and the ORF79 peptide sequence. The primers P1, P2, and P3 were used for RT-PCR to examine the editing of N-atp6 and B-atp6 mRNAs. To determine the 5′ and 3′ ends of the primary (N-atp6) and processed RNA fragments containing B-atp6 or orf79 by a CR-RT-PCR method (Kuhn and Binder, 2002) (see Figure 7B), the primers P4 and P8 were used for the reverse transcription of the circularly ligated RNAs, and the reverse-directed primer pairs P5/P6, P7/P6, and P9/P10 for the PCR. The 5′ and 3′ termini of the processed B-atp6 and orf79 RNA fragments (∼1 and 0.45 kb) are indicated by vertical arrows and open triangles, respectively. The primary 5′ and 3′ termini of N-atp6 and B-atp6/orf79 mRNAs and their different downstream sequences (starting from the bent arrow) have been described (Iwabuchi et al., 1993). The nucleotides identical to cox1 and the encoding amino acids are underlined. The dotted underline indicates a segment identical to the 5′ UTR of a predicted mitochondrial gene orf91 (Itadani et al., 1994). Rectangles indicate the deleted nucleotides in fragment II and the amino acids.
(B) and (C) Effect of orf79 expression with fragments I and II (Figure 1A), indicated by I and II, respectively, on the growth of E. coli cells on an agar plate (B) and in liquid cultures (C) with or without isopropylthio-β-d-galactoside (IPTG). In the liquid cultures (C), IPTG was added when the cell growth reached OD550 = 0.6.
(D) The expressed recombinant ORF79 protein (arrowed) with fragment II.