Figure 1.
cpr22 Is a 3-kb Deletion That Creates a Chimeric ATCNGC-Encoding Gene.
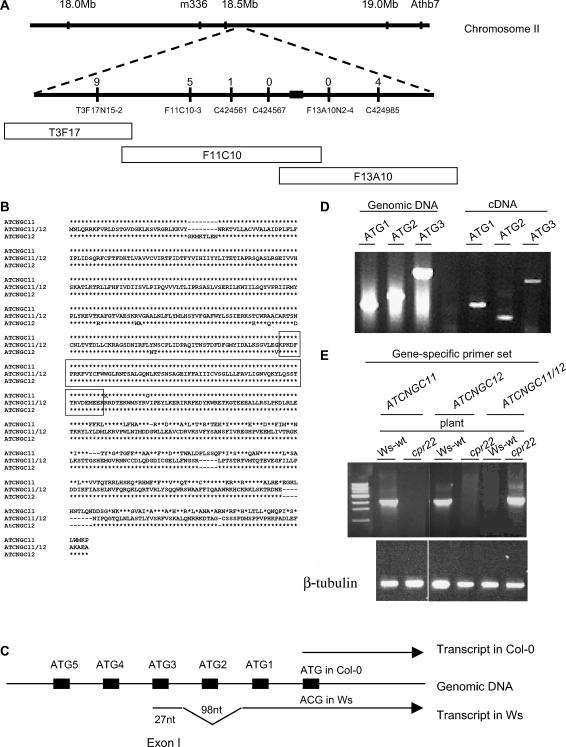
(A) Genetic and physical map of the cpr22 locus on chromosome 2. The top line represents chromosome 2; the bottom line is a larger scale map of the cpr22-containing region, with various markers indicated below the line and the number of recombination events for each marker indicated above it. The location of the 3-kb deletion is indicated as a black box. BAC clones T3F17, F11C10, and F13A10, which span the cpr22-containing region, are represented as open boxes below the map.
(B) Amino acid sequence alignment of ATCNGC11, ATCNGC11/12, and ATCNGC12 from the Ws ecotype. The deduced amino acid sequence for ATCNGC11/12 is in the middle; amino acid residues in ATCNGC11 and/or ATCNGC12 that are identical to those of ATCNGC11/12 are represented as asterisks, whereas residues that differ are indicated above or below the line. The boxed region contains sequences that are identical in all three proteins. Because the predicted ATCNGC11/12 sequence is identical to ATCNGC11 upstream of the boxed sequence and to ATCNGC12 downstream of this sequence, the homologous recombination event that created ATCNGC11/12 likely occurred within this region.
(C) Scheme of the 5′ translated and untranslated regions of the Col-0 and Ws alleles of ATCNGC11. To investigate the position of the translational start codon in the Ws allele of ATCNGC11, RT-PCR analysis was performed with primers corresponding to five ATG codons near the predicted translational start codon of the Col-0 allele. ATG1 to ATG5 are depicted as black boxes on the genomic DNA. This analysis suggested that the transcript for the Ws allele (bottom arrow) contains an extra 98-nucleotide intron and 27-nucleotide exon that are not present in the transcript of the Col-0 allele (top arrow).
(D) PCR analysis of ATCNGC11 (Ws ecotype). Forward primers ATG1, ATG2, and ATG3, which correspond to the first three ATG codons within the predicted 5′ untranslated region of ATCNGC11, were tested for their ability to generate a product using genomic DNA or cDNA from wild-type Ws plants as the template in combination with the ATCNGC11-specific reverse primer UD1R1. ATG1 is closest to the annotated translational start codon, and ATG3 is farther upstream. The products were resolved on an agarose gel and stained with ethidium bromide. See text for additional information.
(E) Semiquantitative RT-PCR analysis of ATCNGC11, ATCNGC12, and ATCNGC11/12 transcript levels in wild-type Ws and cpr22 homozygous plants using gene-specific primers. The products were resolved on an agarose gel and stained with ethidium bromide. β-Tubulin was used as a loading control.