Abstract
The identification of all the substrates of every protein kinase is one of the major challenges of post-genomic research. Here we review a powerful method for tackling this problem that we have developed over the last 5 years. The method has so far been used to identify novel substrates for eight different protein kinases, demonstrating that it is of general utility. Importantly, the method can be used to identify distinct physiological substrates of protein kinases, such as PKB (protein kinase B) and SGK (serum- and glucocorticoid-induced kinase), that are closely related in structure and have similar specificity determinants.
Keywords: glycogen synthase kinase 3 (GSK3), kinase substrate tracking and elucidation (KESTREL), mitogen-activated-protein-kinase-activated protein kinase-2 (MAPKAP-K2), protein kinase B (PKB), serum- and glucocorticoid-induced protein kinase (SGK)
Abbreviations: ARE, AU-rich element; CapZIP, CapZ-interacting protein; CDK, cyclin-dependent protein kinase; CRHSP24, calcium-regulated heatshock protein of 24 kDa; CRMP, collapsin-response mediator protein; EF2, elongation factor 2; EF2K, EF2 kinase; ERK, extracellular-signal-regulated kinase; FLNc, filamin C; GSK3, glycogen synthase kinase 3; HEK, human embryonic kidney; hnRNP, heterogeneous nuclear ribonucleoprotein; HSP27, heat-shock protein of 27 kDa; IGF-1, insulin-like growth factor 1; JNK, c-Jun N-terminal kinase; KSHV, Kaposi sarcoma-associated herpesvirus; KESTREL, kinasesubstrate tracking and elucidation; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; MAPKAP-K2, MAPK-activated protein kinase-2; METTL1, methyltransferase-like protein 1; MKK1, MAPK kinase-1; NDRG, N-myc downstream-regulated gene product; PDK1, 3-phosphoinositide-dependent kinase 1; PIC, pro-inflammatory cytokine; PI3K, phosphoinositide 3-kinase; PKB, protein kinase B; PNGase, peptide N-glycanase; SAPK, stress-activated protein kinase; SAKS1, SAPK-substrate 1; SGK, serum- and glucocorticoid-induced protein kinase; TNF, tumour necrosis factor; UBA, ubiquitin-associated; UBX, ubiquitin-like; VCP, valosin-containing protein
INTRODUCTION
Fifty years after the discovery of the first protein kinase–substrate relationship, namely the activation of glycogen phosphorylase by phosphorylase kinase, only a small fraction of the kinase–substrate pairs estimated to exist have so far been identified. Indeed, the identification of all the substrates of every protein kinase must represent one of the major challenges of post-genomic research. Substrate identification is crucial to gain a detailed molecular understanding of the full repertoire of cellular functions that can be modulated by extracellular signals. It has become obvious that many more powerful methods are going to be needed to address this problem. This review describes one such method, termed KESTREL (kinase substrate tracking and elucidation) that we have introduced and that we and others have exploited over the last 5 years [1].
THE DEVELOPMENT OF KESTREL
In theory, the simplest way to identify the substrates of a protein kinase would be to incubate cell extracts with Mg[γ-32P]ATP and the kinase of interest and to see which proteins in the extract become 32P-labelled. These proteins could then be purified and identified by MS. However, in practice, this approach has rarely been attempted for several reasons. First, cell extracts contain all the protein kinases that are expressed in that cell, as well as their substrates. Thus hundreds of proteins become phosphorylated upon incubation with MgATP and this high ‘background’ phosphorylation would be expected to interfere with the detection of substrates following the exogenous addition of another kinase to the extracts. Secondly, many protein kinases can phosphorylate proteins in vitro that are not their physiological substrates. Therefore proteins identified by such procedures may not turn out to be bona fide substrates. Thirdly, the purification of substrates from a cell extract and their identification can be a difficult and time-consuming process, not to be undertaken without having some confidence that the purified proteins are likely to be physiologically relevant. Fourthly, until recently, and, in the absence of genome sequences, low-abundance proteins were difficult to identify, because methods for their detection were insufficiently sensitive.
We have been able to overcome the above-mentioned problems for detecting and identifying cellular substrates of protein kinases by the introduction of a few simple tricks and by exploiting recent advances in protein analysis. The high background produced by incubation of cell extracts with Mg[γ-32P]ATP can, of course, be reduced by using short incubation times, high concentrations of the added kinase and ATP of very high specific radioactivity. For the last mentioned reason, it is therefore essential to deplete the extracts of ATP and other nucleotides by gel-filtration or dialysis before beginning the analysis. Moreover, subjecting cell extracts to an initial step of ion-exchange chromatography separates endogenous kinases from most of their substrates, and also concentrates the latter, greatly reducing the background problem and increasing the likelihood of detecting substrates that may be missed in total cell extracts [1]. Furthermore, we found empirically that, for many protein kinases, MnATP can be used as the substrate instead of MgATP, which greatly reduces the background noise.
However, perhaps the key innovation that we introduced was to perform the KESTREL screen in parallel using two or three closely related protein kinases whose substrate specificities are very similar towards peptide and/or protein substrates normally used to assay their activities in vitro. The aim of the screen is to detect proteins that are phosphorylated by just one of several closely related protein kinases, which greatly enhances the probability of detecting physiologically relevant substrates [1]. Finally, the development of rapid high-resolution methods for protein purification, coupled with the advent of tryptic mass fingerprinting and tandem-MS microsequencing technology, plus the availability of near-complete mammalian genome sequences, has made the purification and identification of substrates a reasonably rapid and relatively routine procedure.
SUBSTRATE VALIDATION AND CHARACTERIZATION
Once a substrate has been identified, the next steps are to identify the site(s) of phosphorylation, followed by the generation of appropriate phospho-specific antibodies that recognize these sites specifically. An antibody also needs to be generated against the substrate, since the endogenous protein frequently needs to be immunoprecipitated before the phospho-specific antibodies can be used effectively. These antibodies are then exploited to investigate whether the protein becomes phosphorylated at the relevant site(s) in cells stimulated with signals that are known to activate the kinase of interest. To establish whether the protein is phosphorylated in cells by the protein kinase used for the KESTREL screen, or by another protein kinase that is activated in response to the same agonist, it is necessary to demonstrate that the phosphorylation of the endogenous substrate is prevented by relatively specific pharmacological inhibitors of the kinase. It is also necessary to show that phosphorylation does not occur in cells and tissues of mice that do not express the kinase, or in cells from knock-in mice that express a catalytically inactive mutant of the kinase. A summary of the KESTREL procedure and the time it has taken us on average to accomplish each step of the procedure is summarized in Table 1. The following sections provide a brief overview of substrates that have been identified using KESTREL.
Table 1. The KESTREL protocol.
The average time taken for each step is indicated in parentheses.
| 1. | Detect major proteins that are phosphorylated by the kinase of interest that are not phosphorylated by related kinases with similar specificities in vitro (days) |
| 2. | Identify proteins by further purification and tryptic mass fingerprinting (a few weeks) |
| 3. | Show protein is phosphorylated stoichiometrically and at a similar rate to an authentic physiological substrate (less than 1 day) |
| 4. | Identify site of phosphorylation (a few days) |
| 5. | Raise antibodies specific for the phosphorylation site (2–3 months) |
| 6. | Show that the site is phosphorylated in vivo in response to stimuli that are known to activate the kinase of interest. Show that phosphorylation is prevented by drugs that inhibit the activity or activation of the kinase of interest (1 week–2 months) |
| 7. | Show that phosphorylation does not occur in knockout mice lacking the kinase of interest (quick if mice are available!) |
| 8. | Identify the function of the substrate. Then study the effect of phosphorylation on the function of the substrate, e.g. changes in activity, stability, subcellular location and interactions with other proteins (variable) |
VALIDATION OF KESTREL AND ITS EXPLOITATION TO IDENTIFY EF2K (ELONGATION FACTOR 2 KINASE) AS A SUBSTRATE FOR p38δ MAPK (MITOGEN-ACTIVATED PROTEIN KINASE)
Our initial paper validated the KESTREL procedure by using it to identify well-authenticated substrates of several protein kinases [1]. For example, ERKs (extracellular-signal-regulated kinases) 1 and 2, or p38α and p38γ MAPK, were the only substrates detected when muscle extracts were incubated with Mg[γ-32P]ATP and either MKK1 (MAPK kinase-1) or MKK6, respectively. Similarly, MAPKAP-K2 (MAPK-activated protein kinase-2) was detected as a substrate for p38α MAPK [1]. We have also identified MKK1 as a substrate for c-Raf, glycogen synthase as a substrate for GSK3 (glycogen synthase kinase 3), tau as a substrate for CDK5 (cyclin-dependent protein kinase 5) and HSP27 (heat-shock protein of 27 kDa) as a substrate for MAPKAP-K2 in cell extracts (C. Eyers, A. Knebel, C. Morris and P. Cohen, unpublished work), which gave us confidence that the approach was going to identify novel substrates for a variety of protein kinases.
One of the first substrates we discovered with KESTREL was EF2K as a specific substrate for p38δ MAPK. p38δ MAPK, but not the closely related p38α, p38β or p38γ MAPK, phosphorylated EF2K at Ser359, causing its inactivation. Cellular stresses that activate p38δ MAPK triggered the phosphorylation of Ser359 and inactivation of EF2K and, as a consequence, induced the dephosphorylation and activation of its substrate EF2 (elongation factor 2). Thus activation of p38δ MAPK can stimulate the elongation stage of protein synthesis.
Interestingly, we found that IGF-1 (insulin-like growth factor 1) also triggered the phosphorylation of EF2K at Ser359 and the activation of EF2. However, in contrast with stress-induced phosphorylation of Ser359, IGF-1-induced phosphorylation of Ser359 was prevented by prior incubation of the cells with rapamycin. Thus EF2K can be inactivated either by p38δ MAPK or by an as yet unidentified rapamycin-sensitive protein kinase, in response to IGF-1 [1].
IDENTIFICATION OF NEW PHYSIOLOGICAL SUBSTRATES FOR SGK1 (SERUM- AND GLUCOCORTICOID-INDUCED KINASE 1) AND PKB (PROTEIN KINASE B)
SGK1 is the protein kinase that is most similar to PKB (54% identity in the catalytic domain). Both protein kinases are activated by signals that stimulate PI3K (phosphoinositide 3-kinase) and phosphorylate serine and threonine residues that lie in Arg-Xaa-Arg-Xaa-Xaa-Ser/Thr motifs. Overexpression studies performed in other laboratories suggested that these protein kinases may have several physiological substrates in common, such as GSK3 [2,3] and FOXO3a [4]. However, it was shown subsequently that the IGF-1-induced phosphorylation of these proteins was not impaired in embryonic stem cells that express PDK1[L155E] (the L155E mutant of 3-phosphoinositide-dependent kinase 1), which activates PKB normally, but cannot activate SGK1 [5]. Moreover, other laboratory groups have shown that the phenotypes of PKB- and SGK1-knockout mice are quite different. For example, mice that do not express the PKBβ isoform have impaired insulin-stimulated glucose uptake and become diabetic as they age [6], while SGK1-deficient mice have impaired ability to adequately decrease Na+ ion excretion when dietary NaCl is restricted [7]. By carrying out KESTREL screens with both protein kinases in parallel, we were able to identify proteins that are phosphorylated by SGK and not PKB, and vice versa.
Identification of NDRG1 (N-myc downstream-regulated gene product 1) and NDRG2 as physiological substrates for SGK1
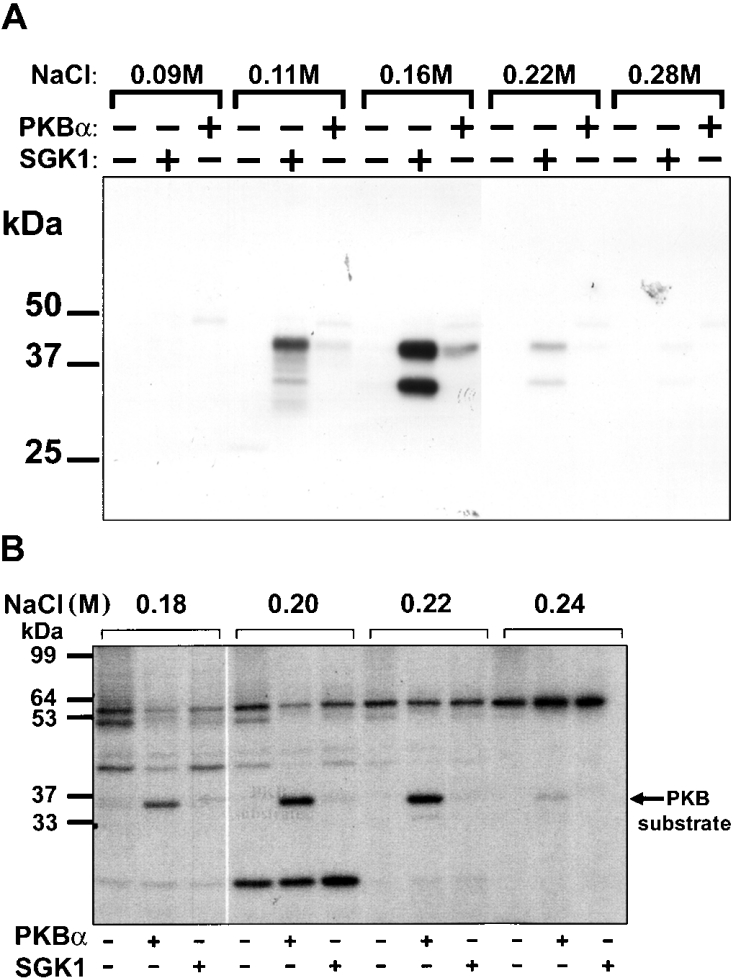
We used KESTREL to identify NDRG1 and NDRG2 as proteins that are phosphorylated by SGK, but not by PKB, in vitro (Figure 1A). Through the use of SGK1-deficient cells and siRNA (small interfering RNA) technology, we were able to establish that both proteins are phosphorylated by SGK1 in cells, one of the first physiological substrates of this protein kinase to be identified [8]. We found that SGK1 phosphorylates NDRG2 at three sites and NDRG1 at five sites, three of which, Thr346, Thr356 and Thr366, are located in identical nonapeptide sequences that are repeated three times near the C-terminus of NDRG1. These residues in NDRG1 and NDRG2 are not phosphorylated in tissues from SGK1−/− mice. Interestingly, phosphorylation by SGK1 primes NDRG1 for phosphorylation by GSK3 at Ser342, Ser352 and Ser362 respectively. These three sites are also phosphorylated in HeLa cells, and their phosphorylation is suppressed by pharmacological inhibition of GSK3 or in SGK1-deficient cells. Thus the C-terminus of NDRG1 is phosphorylated at six positions by the combined actions of SGK1 and GSK3 [8].
Figure 1. Detection of two proteins phosphorylated relatively specifically in vitro by SGK1 and PKBα respectively.
(A) Identification of a protein phosphorylated much more rapidly by SGK1 than PKB. The proteins not retained on heparin–Sepharose were chromatographed on Source 15 Q-Sepharose. Each fraction was incubated for 4 min at 30 °C with 2 mM MgCl2/20 nM [γ-32P]ATP in the absence (−) or presence (+) of 0.3 unit/ml SGK1 or PKBα, denatured in SDS, subjected to SDS/PAGE, transferred to Immobilon P membranes and autoradiographed. Two substrates for SGK1 with apparent molecular masses of 45 kDa and 35 kDa, eluting between 0.11 M and 0.22 M NaCl, were detected by autoradiography. Reproduced from [8] with permission. © 2004 The Biochemical Society. (B) Identification of a 36 kDa protein phosphorylated by PKB, but not by SGK, after chromatography on a MonoQ column. Detection of a 36 kDa substrate for PKBα in HeLa extracts. HeLa cell extracts were fractionated from 0–5% (w/v) PEG 6000, the 5% supernatant was desalted and the material (950 mg of protein) was chromatographed on a 20 ml column of Source Q. Aliquots of each fraction were incubated for 4 min at 30 °C as described previously [1] at a further 10-fold dilution in the presence of 50 mM Tris/HCl, pH 7.5, 1 mM EGTA, 0.1% (v/v) 2-mercaptoethanol, 10 mM MgCl2 and 20 nM [γ-32P]ATP (4×106 c.p.m./pmol) in the presence (+) or absence (−) of PKBα or SGK1, each at 0.4 unit/ml [30]; 1 unit is the amount that catalyses the phosphorylation of 1 nmol of substrate peptide in 1 min. After SDS/PAGE and autoradiography, a 36 kDa protein eluting at approx. 0.2 M NaCl was detected that was phosphorylated by PKBα, but very poorly by SGK1. Autophosphorylation of PKB and SGK1 was negligible under the conditions used. Reprinted by permission from Macmillan Publishers Ltd: EMBO Journal [10], © 2005.
The nonapeptide repeat present in NDRG1 lies in a canonical Arg-Xaa-Arg-Xaa-Xaa-Ser/Thr sequence, which is normally phosphorylated by either SGK or PKB. Interestingly, we found that a synthetic peptide corresponding to the nonapeptide repeat is phosphorylated efficiently by SGK, but extremely poorly by PKB. This allowed us to identify three residues in this sequence at positions n+1, n+2 and n−4 (where n is the site of phosphorylation) that are negative determinants for phosphorylation by PKB, but much less so for SGK1. The mutation of these three residues to other amino acid side chains transforms the nonapeptide into an excellent substrate for either PKB or SGK1 [9]. These findings may facilitate the identification of further SGK-specific substrates.
NDRG1 was originally identified by other laboratory groups as a protein which is up-regulated by a variety of stress signals, p53 expression and DNA damage, and its expression is inhibited under conditions of cell growth. It was also identified as a protein that is up-regulated in mouse embryos that are deficient in N-myc and down-regulated in tumours. Its expression is induced by stimuli that promote differentiation in cancer cells, and it has been reported to be a metastasis-suppressor gene. A mutation in NDRG1 is the cause of Charcot–Marie–Tooth disease type 4D, a hereditary disease that is characterized by muscle weakness, sensory loss and neural deafness: symptoms caused by demyelination of peripheral nerves. There is Schwann cell dysfunction in NDRG1-deficient mice, suggesting that NDRG1 is essential for maintenance of the myelin sheaths in the peripheral nerves. NDRG2 has been reported to be induced by mineralocorticoid hormones, such as aldosterone, which also induce the expression of SGK1 (reviewed in [8]). It will clearly be of considerable interest to understand how the phosphorylation of NDRG1 and NDRG2 regulates the functions of these interesting proteins.
METTL1 (methyltransferase-like protein 1) as a physiological substrate for PKB
We used KESTREL to identify METTL1 as a protein that is phosphorylated efficiently by PKB, but very poorly by SGK (Figure 1A). PKB phosphorylated METTL1 at Ser27, and this residue became phosphorylated in IGF-1-stimulated cells. The IGF-1-induced phosphorylation was prevented by inhibition of PI3K, did not occur in cells deficient in PDK1 (the ‘upstream’ activator of PKB), but occurred normally in cells that express the PDK1[L155E] mutant instead of the wild-type protein; this mutant activates PKB, but not SGK [5]. These results establish METTL1 as a new physiological substrate for PKB [10].
METTL1 was shown by Eric Phizicky and colleagues to catalyse the 7-methylation of a guanosine residue found at position 46 in many tRNAs [11]. We found that the PKB-catalysed phosphorylation of METTL1 at Ser27 inactivated this enzyme [10]. The mutation of Ser27 to Asp or Glu, to mimic the effect of phosphorylation by introducing a negative charge, also inactivated METTL1. Disruption of the Saccharomyces cerevisiae orthologue of METTL1 produced a yeast strain that could not grow on minimal media, but grew normally on glucose-containing media. This defect was suppressed by the co-expression of human METTL1 and its regulatory subunit WDR4, but not by expression of the METTL1[S27D] or METTL1[S27E] mutants. Thus the introduction of a negative charge at position 27, whether by phosphorylation or mutation, produces an enzyme that is inactive in cells [10].
Remarkably, the sequence surrounding Ser27 (RQRAHSNP) is perfectly conserved from yeast to humans, indicating that this region of METTL1 is critical for function and that the inactivation of METTL1 by phosphorylation is a control mechanism that is likely to operate in all eukaryotes.
The studies that we have carried out so far suggest that, under the adverse conditions of nutrient deprivation, the m7G46 modification of tRNA is important to maintain a minimum rate of protein synthesis, but, when the level of insulin is elevated and the availability of amino acids and glucose is high, the m7G46 modification is benign. It will clearly be important in the future to delineate the precise role of this tRNA modification before the full physiological significance of these findings can be understood.
Identification of FLNc (filamin C) and CRHSP24 (calcium-regulated heat-shock protein of 24 kDa) as new physiological substrates for PKB
We also identified FLNc [12] and CRHSP24 [13] as proteins in cell extracts that are phosphorylated efficiently by PKB, but not by SGK, in vitro, and went on to establish that they are physiological substrates for PKB using a similar methodology to that described above for METTL1 [10].
FLNc, a muscle-specific splice variant of the actin-binding protein filamin, contains a 78-amino-acid-residue insert located between the 19th and 20th immunoglobulin-like repeats, of which 24 are present in all filamin variants. Strikingly, it is within this insert region unique to FLNc that the residue phosphorylated by PKB (Ser2213) is situated [12]. In skeletal muscle, FLNc is known to associate with the Z-disc of the myofibrillar apparatus and to bind directly to the Z-disc proteins FATZ (filamin-, actinin- and telethonin-binding protein of the Z-disc) and myotilin. It also interacts with sarcoglycans α and β at the sarcolemma, which are components of the dystrophin–dystroglycan complex (reviewed in [12]). It will be interesting to investigate whether phosphorylation by PKB affects the interaction of FLNc with these proteins.
We found that CRHSP24 is phosphorylated at Ser52 by PKB in response to signals that activate PI3K [13], and, interestingly, this site is located just proximal to the cold-shock domain that comprises 40% of this protein. CRHSP24 was originally identified by others as a protein that underwent dephosphorylation by the calcium/calmodulin-dependent protein phosphatase 2B (also called calcineurin) in cholecystokinin-stimulated pancreatic acinar cells [14,15]. However, dephosphorylation was only monitored by a change in the electrophoretic migration of CRHSP24, and the phosphorylated residue(s) was (were) not identified. Interestingly, the phosphorylation of CRHSP24 at Ser52 does not decrease in response to the calcium ionophore ionomycin in HEK-293 (human embryonic kidney) cells, suggesting that calcineurin may dephosphorylate another site(s) on the protein [13].
IDENTIFICATION OF CALDESMON AS A SUBSTRATE FOR K-CYCLIN–CDK6
KSHV (Kaposi sarcoma-associated herpesvirus) encodes a D-like cyclin (K-cyclin), which is thought to contribute to the oncogenicity of the virus. K-cyclin activates cellular CDK6, generating an enzyme which is thought to have a substrate selectivity deviant from CDK6 or CDK4, which are normally activated by cellular D-cyclins. KESTREL has been used to identify caldesmon as a novel substrate for K-cyclin–CDK6 complexes that is not phosphorylated by D-cyclin–CDK6 [16].
Caldesmon is known to play a central role in regulating microfilament organization, by controlling cell shape, adhesion, cytokinesis and motility. K-cyclin–CDK6 phosphorylated four Ser-Pro/Thr-Pro sequences near the C-terminus of caldesmon, abolishing its ability to bind to its effector protein actin and its regulator protein calmodulin. Moreover, caldesmon became hyperphosphorylated in cells following K-cyclin expression and in KSHV-transformed lymphoma cells. Furthermore, expression of exogenous K-cyclin resulted in microfilament loss and changes in cell morphology that were dependent on CDK activity and were reversed by expression of a caldesmon mutant that could not be phosphorylated. Taken together, these data strongly suggest that K-cyclin expression induces the phosphorylation of caldesmon, modulates its activity in cells and through this regulates microfilament function. These findings establish a novel link between KSHV infection and the regulation of the actin cytoskeleton, which is likely to contribute to the oncogenicity of the virus [16].
IDENTIFICATION OF THE CRMPs (COLLAPSIN-RESPONSE MEDIATOR PROTEINS) AS NEW PHYSIOLOGICAL SUBSTRATES FOR GSK3
A KESTREL screen of a brain extract led to the identification of CRMP2 and CRMP4 as physiological substrates for GSK3 [17]. Expression of wild-type CRMP2 in primary hippocampal neurons of SH-SY5Y neuroblastoma cells promoted axon elongation. However, a CRMP2 mutant that cannot be phosphorylated by GSK3 has a greatly reduced ability to promote axon elongation, similar to observations made after pharmacological inhibition of GSK3. These observations suggest that the phosphorylation of CRMP2 by GSK3 alters the ability of CRMP2 to promote axon elongation [17]. Interestingly, the amino acid residues targeted by GSK3 comprise a hyperphosphorylated epitope, first identified in plaques isolated from the brains of patients with Alzheimer's disease [18]. GSK3 has also been implicated in the hyperphosphorylation of other proteins that are enriched in plaques, such as the amyloid precursor protein [19] and the neurofilament L, M and H proteins [20].
IDENTIFICATION OF SUBSTRATES FOR SAPKs (STRESS-ACTIVATED PROTEIN KINASES)
Identification of hnRNP (heterogeneous nuclear ribonucleoprotein) A0 as a new physiological substrate for MAPKAP-K2
Bacterial infection triggers the production of PICs (pro-inflammatory cytokines), such as TNF (tumour necrosis factor), which then mount the immune responses that fight and destroy the invading pathogen. Stimulation of PIC production is mediated by stabilization and/or increased translation of the mRNAs encoding PICs and involves neutralization of the destabilizing effects of the AREs (AU-rich elements) that are present in the 3′ non-coding region of these mRNAs. The signalling pathway leading to activation of MAPKAP-K2 is required for the LPS (lipopolysaccharide)-induced production of several PICs, because drugs that inhibit p38α MAPK suppress the production of TNF [21], and there is an 85–90% reduction in LPS-induced production of TNF, interleukin 6 and interferon γ in mice that are deficient in MAPKAP-K2 [22]. We reasoned that MAPKAP-K2 may exert its effects on PIC production by phosphorylating proteins that are associated with the ARE and therefore purified this fraction by affinity chromatography, before carrying out a KESTREL analysis. This led to the identification of hnRNP A0 as a new physiological substrate for MAPKAP-K2 [23]. LPS stimulation induced the binding of hnRNP A0 to the mRNAs encoding TNF and cyclo-oxygenase 2 and, most strikingly, to MIP2 (macrophage inflammatory protein 2). hnRNP A0 is therefore a candidate to be involved in mediating the stabilization/translation of these PICs [23].
Identification of CapZIP (CapZ-interacting protein) as a substrate for several SAPKs
A protein that is only expressed in muscle and in cells of the immune system was identified in three separate KESTREL screens using MAPKAP-K2, p38γ MAPK or p38δ MAPK [24]. It was found to bind specifically to CapZ, an actin-capping protein, and was therefore termed CapZIP.
MAPKAP-K2 phosphorylated CapZIP at Ser179 in vitro and in cells exposed to stress stimuli. The anisomycin-induced phosphorylation of Ser179 was prevented by SB 203580, consistent with phosphorylation by MAPKAP-K2 and/or the closely related isoform MAPKAP-K3. In contrast, osmotic-shock-induced phosphorylation was not prevented by SB 203580, indicating that at least one other SB 203580-insensitive protein kinase can phosphorylate this site in cells.
p38γ MAPK or p38δ MAPK or JNK1 (c-Jun N-terminal kinase 1) phosphorylated CapZIP at multiple sites, four of which were identified. One of these (Ser108) was studied in detail and was shown to become phosphorylated in cells that were exposed to anisomycin or osmotic shock. However, the relevant protein kinase(s) is unclear, because the osmotic-shock-induced phosphorylation of Ser108 occurred normally in splenocytes from mice that do not express either p38γ MAPK or p38δ MAPK. Also, the phosphorylation was not suppressed by SB 203580 or by pharmacological inhibition of the classical MAPK cascade, which excludes the involvement of SAPK2a/p38α, SAPK2b/p38β, ERK1 or ERK2. The relevant protein kinase(s) could therefore be one or more JNK isoforms, since CapZIP is a remarkably good substrate for JNK1 in vitro [24]. Alternatively, CapZIP may be targeted by several SAPKs.
Importantly, stimuli that induce the phosphorylation of CapZIP trigger its dissociation from CapZ [24]. CapZ is a heterodimer composed of two subunits, CapZα and CapZβ. It is thought to regulate actin filament assembly and organization by binding to the barbed ends of the filaments. Here it works as a ‘cap’, preventing the addition and/or the loss of actin monomers at the ends, and can therefore reduce the length of actin filaments. One could therefore envisage that the interaction between CapZIP and CapZ affects the ability of the latter to remodel actin filaments. Such an effect is presumably lost when CapZIP is phosphorylated and dissociates from CapZ.
HSP27, a well-established substrate for MAPKAP-K2, is known to act as an actin cap-binding protein when phosphorylated, and the phosphorylation of this protein appears to contribute to enhanced actin polymerization and reorganization in vivo, a process thought to be involved in protecting the cytoskeleton from stress-induced damage, thereby aiding cell survival [25]. Interestingly, it was reported recently that the amount of HSP27 and CapZ associated with actin increased and decreased respectively when smooth muscle from the saphenous vein was exposed to haemodynamic stress [26]. Both of these changes would favour the generation of contractile stress fibres.
SAKS1 (SAPK-substrate 1)
SAKS1 is another protein that was identified by a KESTREL screen using p38γ MAPK [27]. It was found to be phosphorylated at Ser200 in vitro not only by p38γ MAPK, but also by p38δ MAPK and JNK1, and this residue became phosphorylated when HEK-293 cells were exposed to cellular stresses, such as osmotic shock. However, as for CapZ, the SAPK(s) which phosphorylates SAKS1 in vivo has yet to be identified because phosphorylation occurred normally in cells that do not express either p38γ MAPK or p38δ MAPK, and nor was phosphorylation suppressed by SB 203580 or pharmacological inhibition of the classical MAPK cascade.
SAKS1 contains an N-terminal UBA (ubiquitin-associated) domain and a C-terminal UBX (ubiquitin-like) domain. The UBA domain binds polyubiquitin, while the UBX domain interacts specifically with VCP (valosin-containing protein), a member of the AAA (ATPases associated with various cellular activities) family of ATPases. VCP has been implicated in many cell functions, including the ATP-dependent unfolding of polyubiquitinated proteins. VCP can bind simultaneously to SAKS1 and PNGase (peptide N-glycanase), an enzyme that removes high-mannose-containing oligosaccharides from misfolded glycoproteins. SAKS1, VCP and PNGase can be immunoprecipitated from cell extracts with antibodies against S5a, a component of the 19 S proteasome that binds to polyubiquitinated proteins. These observations suggest that SAKS1 is an adaptor protein that directs VCP to polyubiquitinated proteins and PNGase to misfolded glycoproteins, facilitating their unfolding, deglycosylation and destruction by the proteasome [27]. The phosphorylation of SAKS1 at Ser200 is presumed to regulate the function of SAKS1 in a way that is not yet understood.
THE KESTREL METHOD — A SUMMARY
Over the last 5 years, we and others have demonstrated that KESTREL is a powerful and rapid method for identifying new physiological substrates of protein kinases (Table 1). The method requires active purified kinase to be available and at least 500 mg of cell extract protein. This quantity of cell extract is necessary to enable low-abundance proteins to be isolated in the microgram amounts needed for their identification and phosphorylation site analysis. The KESTREL method can be used for nearly all protein kinases, apart from the very few that have an exceptionally high Km for ATP (>0.15 mM). In practice, we have found that the activities of these protein kinases are so low at the concentration of ATP used for the initial KESTREL screen (20 nM) that their substrates are not detected in cell extracts. Using the KESTREL method, we and others have identified new substrates for eight different protein kinases (Table 2). The method has been exploited to identify new substrates for SAPK4/p38δ [1], SGK1 [8], PKB [10,12,13], K-cyclin–CDK6 [16], GSK3 [17], MAPKAP-K2 [23,24], Aurora A [28] and SAPKs [24,27], demonstrating that it is of wide application. Even when the substrate has turned out not to be phosphorylated by the protein kinase used for the initial KESTREL screen, it has usually been shown to undergo phosphorylation at the relevant site(s) in cells by a related protein kinase [24,27] or another protein kinase [29].
Table 2. Novel physiological substrates for eight different protein kinases identified by the use of KESTREL.
The major frustration encountered using KESTREL, in common with any other methods used to screen for novel substrates of protein kinases, is that many of the new substrates identified have turned out to be proteins that had either never been studied previously or whose functions were largely unknown (e.g. CapZIP, SAKS1, NDRG1/2, CRHSP24 and FLNc). Therefore, even though it was relatively easy to establish that these proteins were physiological substrates for particular protein kinases, a huge amount of effort was still required to obtain information about their functions, before the role of phosphorylation could be addressed. Nevertheless, a huge number of interesting projects have been opened up by the studies that have been undertaken, whose full biological significance will be revealed more fully in the years to come.
Acknowledgments
We thank the past and present members of our laboratory who have been involved in validating the KESTREL method over the last 5 years, namely Gillian Auld, David Campbell, Robert Cartlidge, Claire Eyers, Helen McNeill, Nick Morrice, James Murray and Simon Rousseau, and the MRC and Royal Society for financial support. The KESTREL method was developed while A.K. was the recipient of an EMBO Long Term Fellowship. A.K. has set up a company that offers KESTREL as a service for those who want to use it without setting it up ‘in house’ (http://www.kinasource.co.uk).
References
- 1.Knebel A., Morrice N., Cohen P. A novel method to identify protein kinase substrates: eEF2 kinase is phosphorylated and inhibited by SAPK4/p38δ. EMBO J. 2001;20:4360–4369. doi: 10.1093/emboj/20.16.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kobayashi T., Cohen P. Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem. J. 1999;339:319–328. [PMC free article] [PubMed] [Google Scholar]
- 3.Park J., Leong M. L., Buse P., Maiyar A. C., Firestone G. L., Hemmings B. A. Serum and glucocorticoid-inducible kinase (SGK) is a target of the PI 3-kinase-stimulated signaling pathway. EMBO J. 1999;18:3024–3033. doi: 10.1093/emboj/18.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunet A., Park J., Tran H., Hu L. S., Hemmings B. A., Greenberg M. E. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Mol. Cell. Biol. 2001;21:952–965. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins B. J., Deak M., Arthur J. S. C., Armit L. J., Alessi D. R. In vivo role of the PIF-binding docking site of PDK1 defined by knock-in mutation. EMBO J. 2003;22:4202–4211. doi: 10.1093/emboj/cdg407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bae S. S., Cho H., Mu J., Birnbaum M. J. Isoform-specific regulation of insulin-dependent glucose uptake by Akt/protein kinase B. J. Biol. Chem. 2003;278:49530–49536. doi: 10.1074/jbc.M306782200. [DOI] [PubMed] [Google Scholar]
- 7.Wulff P., Vallon V., Huang D. Y., Volkl H., Yu F., Richter K., Jansen M., Schlunz M., Klingel K., Loffing J., et al. Impaired renal Na+ retention in the sgk1-knockout mouse. J. Clin. Invest. 2002;110:1263–1268. doi: 10.1172/JCI15696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray J. T., Campbell D. G., Morrice N., Auld G. C., Shpiro N., Marquez R., Peggie M., Bain J., Bloomberg G. B., Grahammer F., et al. Exploitation of KESTREL to identify N-myc downstream-regulated gene family members as physiological substrates for SGK1 and GSK3. Biochem J. 2004;384:477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray J. T., Cummings L. A., Bloomberg G. B., Cohen P. Identification of different specificity requirements between SGK1 and PKBα. FEBS Lett. 2005;579:991–994. doi: 10.1016/j.febslet.2004.12.069. [DOI] [PubMed] [Google Scholar]
- 10.Cartlidge R. A., Knebel A., Peggie M., Alexandrov A., Phizicky E. M., Cohen P. The tRNA methylase METTL1 is phosphorylated and inactivated by PKB and RSK in vitro and in cells. EMBO J. 2005;24:1696–1705. doi: 10.1038/sj.emboj.7600648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexandrov A., Martzen M. R., Phizicky E. M. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA. 2002;8:1253–1266. doi: 10.1017/s1355838202024019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray J. T., Campbell D. G., Peggie M., Mora A., Cohen P. Identification of filamin C as a new physiological substrate for PKBα using KESTREL. Biochem. J. 2005;384:489–494. doi: 10.1042/BJ20041058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Auld G. C., Campbell D. G., Morrice N., Cohen P. Identification of calcium-regulated heat stable protein of 24 kDa (CRHSP24) as a physiological substrate for PKB and RSK using KESTREL. Biochem. J. 2005;389:775–783. doi: 10.1042/BJ20050733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groblewski G. E., Yoshida M., Bragado M. J., Ernst S. A., Leykam J., Williams J. A. Purification and characterization of a novel physiological substrate for calcineurin in mammalian cells. J. Biol. Chem. 1998;273:22738–22744. doi: 10.1074/jbc.273.35.22738. [DOI] [PubMed] [Google Scholar]
- 15.Schafer C., Steffen H., Krykowski K. J., Goke B., Groblewski G. E. CRHSP-24 phosphorylation is regulated by multiple signalling pathways in pancreatic acinar cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;285:G726–G734. doi: 10.1152/ajpgi.00111.2003. [DOI] [PubMed] [Google Scholar]
- 16.Cuomo M. E., Knebel A., Platt G., Morrice N., Cohen P., Mittnacht S. Regulation of microfilament organization by Kaposi sarcoma-associated herpes virus-cyclin·CDK6 phosphorylation of caldesmon. J. Biol. Chem. 2005;280:35844–35858. doi: 10.1074/jbc.M503877200. [DOI] [PubMed] [Google Scholar]
- 17.Cole A. R., Knebel A., Morrice N., Robertson L. A., Irving A. J., Connolly C. N., Sutherland C. GSK-3 phosphorylation of the Alzheimer epitope within collapsin response mediator proteins regulates axon elongation in primary neurons. J. Biol. Chem. 2004;279:50176–50180. doi: 10.1074/jbc.C400412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu Y., Hamajima N., Ihara Y. Neurofibrillary tangle-associated collapsin response mediator protein-2 (CRMP-2) is highly phosphorylated on Thr-509, Ser-518 and Ser-522. Biochemistry. 2000;39:4267–4275. doi: 10.1021/bi992323h. [DOI] [PubMed] [Google Scholar]
- 19.Aplin A. E., Gibb G. M., Jacobsen J. S., Gallo J. M., Anderton B. H. In vitro phosphorylation of the cytoplasmic domain of the amyloid precursor protein by glycogen synthase kinase-3β. J. Neurochem. 1996;67:699–707. doi: 10.1046/j.1471-4159.1996.67020699.x. [DOI] [PubMed] [Google Scholar]
- 20.Guan R. J., Khatra B. S., Cohlberg J. A. Phosphorylation of bovine neurofilament proteins by protein kinase FA (glycogen synthase kinase 3) J. Biol. Chem. 1991;266:8262–8267. [PubMed] [Google Scholar]
- 21.Lee J. C., Laydon J. T., McDonnell P. C., Gallagher T. F., Kumar S., Green D., McNulty D., Blumenthal M. J., Heys J. R., Landvatter S. W., et al. A protein kinase involved in the regulation of proinflammatory cytokine synthesis. Nature (London) 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 22.Koylyarov A., Neininger A., Schubert C., Eckert R., Birchmeier C., Volk H. D., Gaestel M. MAPKAP-K2 is essential for LPS-induced TNFα biosynthesis. Nat. Cell Biol. 1999;1:94–97. doi: 10.1038/10061. [DOI] [PubMed] [Google Scholar]
- 23.Rousseau S., Morrice N., Peggie M., Campbell D. G., Gaestel M., Cohen P. Inhibition of SAPK2a/p38 prevents hnRNP A0 phosphorylation by MAPKAP-K2 and its interaction with cytokine mRNAs. EMBO J. 2002;21:6505–6514. doi: 10.1093/emboj/cdf639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eyers C. E., McNeil H., Knebel A., Morrice N., Arthur S. J. C., Cuenda A., Cohen P. the phosphorylation of CapZ-interacting protein (CapZIP) by stress-activated protein kinases triggers its dissociation from CapZ. Biochem. J. 2005;389:127–135. doi: 10.1042/BJ20050387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guay J., Lambert H., Gingras-Breton G., Lavioe J. N., Landry J. Regulation of actin filament dynamics by p38 map kinase-mediated phosphorylation of heat shock protein 27. J. Cell Sci. 1997;110:357–368. doi: 10.1242/jcs.110.3.357. [DOI] [PubMed] [Google Scholar]
- 26.McGregor E., Kempster L., Wait R., Gosling M., Dunn M. J., Powell J. T. F-actin capping (CapZ) and other contractile saphenous vein smooth muscle proteins are altered by hemodynamic stress: a proteonomic approach. Mol. Cell. Proteomics. 2004;3:115–124. doi: 10.1074/mcp.M300046-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.McNeil H., Knebel A., Arthur J. S. C., Cuenda A., Cohen P. A novel UBA and UBX domain protein that binds polyubiquitin and VCP and is a substrate for SAPKs. Biochem. J. 2004;384:391–400. doi: 10.1042/BJ20041498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Troiani S., Uggeri M., Moll J., Isacchi A., Kalisz H. M., Rusconi L., Valsasina B. Searching for biomarkers of Aurora-A kinase activity: identification of in vitro substrates through a modified KESTREL approach. J. Proteome Res. 2005;4:1296–1303. doi: 10.1021/pr050018e. [DOI] [PubMed] [Google Scholar]
- 29.Haydon C. E., Watt P. W., Morrice N., Knebel A., Gaestel M., Cohen P. Identification of a phosphorylation site on skeletal muscle myosin light chain kinase that becomes phosphorylated during muscle contraction. Arch. Biochem. Biophys. 2002;397:224–231. doi: 10.1006/abbi.2001.2625. [DOI] [PubMed] [Google Scholar]
- 30.Davies S. P., Reddy H., Caivano M., Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]